Cytoscape Automation in R using Rcy3
Ruth Isserlin
July 26, 2018
BioC 2018 - Toronto, Ontario

Overview
- Ruth Isserlin - ruth dot isserlin (at) utoronto (dot) ca
- Brendan Innes - brendan (dot) innes (at) mail (dot) utoronto (dot) ca
- Jeff Wong - jvwong (at) gmail (dot) com
- Gary Bader - gary (dot) bader (at) utoronto (dot) ca
Download and Install:
- Slides - https://cytoscape.github.io/cytoscape-tutorials/presentations/bioc2018_Rcy3_intro#/
- Cytoscape - http://www.cytoscape.org/download.php
- RNotebook files - https://github.com/Bioconductor/BiocWorkshops/blob/master/230_Isserlin_RCy3_intro.Rmd
- Workshop Book - https://bioconductor.github.io/BiocWorkshops/BioC2018.pdf
- R packages (list below)
- Cytoscape Apps (listed below) - in Cytoscape go to Apps->App Manager to install apps.
| in RStudio install the following packages |
|---|
| RCy3 |
| gProfileR |
| RCurl |
| EnrichmentBrowser |
| in Cytoscape install |
|---|
| EnrichmentMap pipeline Collection |
| Functional Enrichment Collection |
| aMatReader |
Prerequisites
| Workshop prerequisites |
|---|
| Basic knowledge of R syntax |
| Basic knowledge of Cytoscape software |
| Familiarity with network biology concepts |
| R / Bioconductor packages |
|---|
| RCy3 |
| gProfileR |
| RCurl |
| EnrichmentBrowser |
Outline
| Activity | Time |
|---|---|
| Introduction | 15m |
| Driving Cytoscape from R | 15m |
| Creating, retrieving and manipulating networks | 15m |
| Summary | 10m |
Workshop goals and objectives
Learning goals - Know when and how to use Cytoscape in your research area
- Generalize network analysis methods to multiple problem domains
- Integrate Cytoscape into your bioinformatics pipelines
Learning objectives - Programmatic control over Cytoscape from R
- Publish, share and export networks
Background
Cytoscape

- Cytoscape(Shannon et al.) is a freely available open-source, cross platform network analysis software.
- Cytoscape(Shannon et al.) can visualize complex networks and help integrate them with any other data type.
- Cytoscape(Shannon et al.) has an active developer and user base with more than 300 community created apps available from the (Cytoscape App Store)[apps.cytoscape.org].
- Check out some of the tasks you can do with Cytoscape in our online tutorial guide - tutorials.cytoscape.org
Cytoscape Apps

Overview of network biology themes and concepts
Why Networks?
Networks are everywhere… - Molecular Networks
- Cell-Cell communication Networks
- Computer networks
- Social Networks
- Internet
Networks are powerful tools… - Reduce complexity
- More efficient than tables
- Great for data integration
- Intuitive visualization
Often data in our pipelines are represented as data.frames, tables, matrices, vectors or lists. Sometimes we represent this data as heatmaps, or plots in efforts to summarize the results visually. Network visualization offers an additional method that readily incorporates many different data types and variables into a single picture.
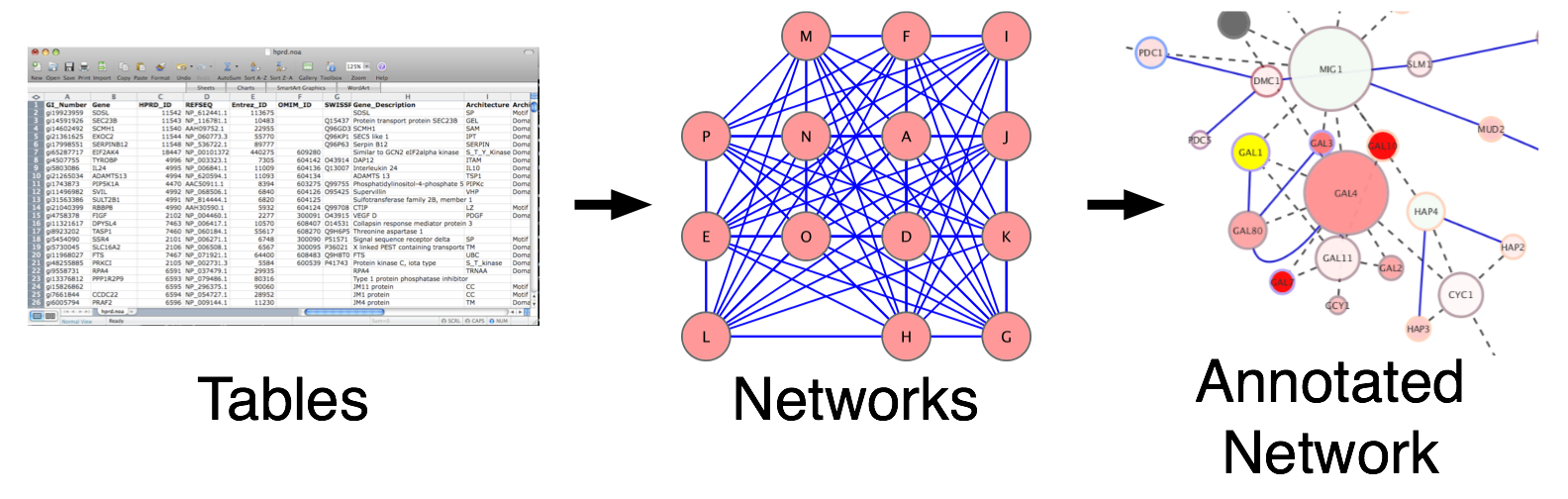
Network types
In order to translate your data into a network it is important to define the entities and their relationships. Entities and relationships can be anything. They can be user defined or they can be queried from a database.
Examples of Networks and their associated entities: - Protein - Protein interaction network
- Gene - gene interaction network -
- Coexpression network
- Enrichment Map
- Social network
- Copublication network
Networks as Tools
Networks can be used for two main purposes but often go hand in hand.
Analysis - Topological properties
- Hubs and subnetworks
- Cluster, classify, and diffuse
- Data integration
Visualization - Data overlays
- Layouts and animation
- Exploratory analysis
- Context and interpretation
Translating biological data into Cytoscape using RCy3
Networks offer us a useful way to represent our biological data. But how do we seamlessly translate our data from R into Cytoscape?
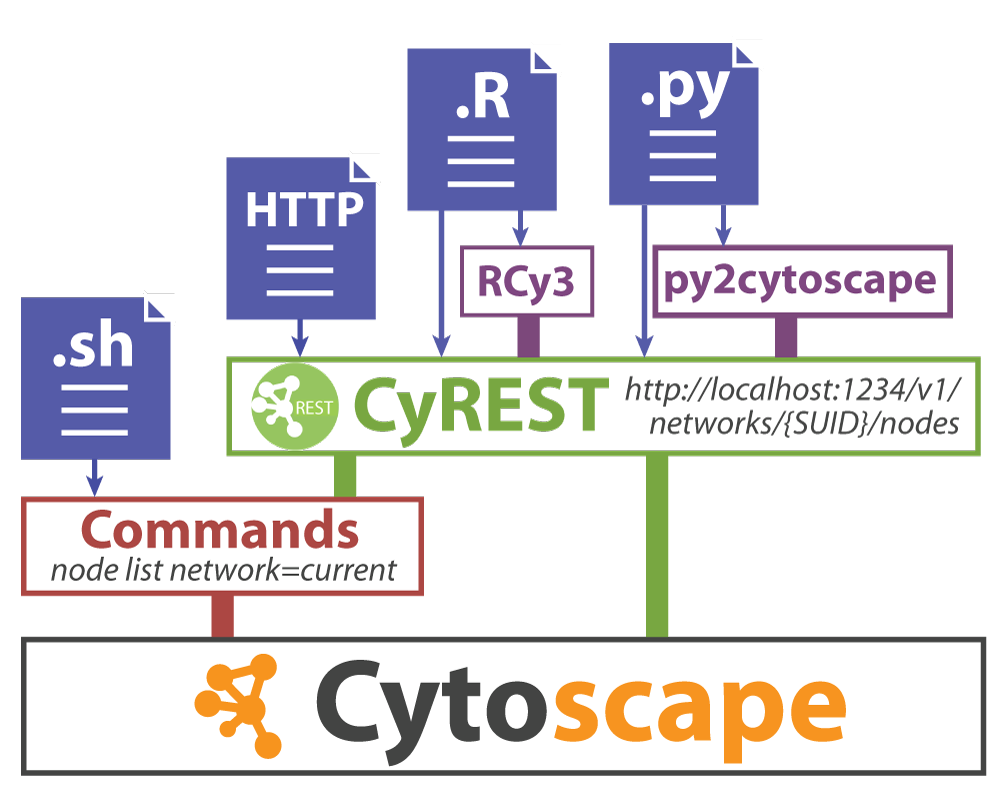
Set up
In order to create networks in Cytoscape from R you need:
- RCy3 - a biocondutor package
if(!"RCy3" %in% installed.packages()){
install.packages("BiocManager")
BiocManager::install("RCy3")
}
library(RCy3)
Set up - (part 2)
- Cytoscape - Download and install Cytoscape 3.6.1. or higher. Java 9 in not supported. Please make sure that Java 8 is installed.
- Install additional cytoscape apps that will be used in this workshop. If using cytoscape 3.6.1 or older the apps need to manually installed through the app manager in Cytoscape or through your web browser. (click on the method to see detailed instructions)
- Functional Enrichment Collection -a collection of apps to retrieve networks and pathways, integrate and explore the data, perform functional enrichment analysis, and interpret and display your results.
- EnrichmentMap Pipeline Collection - a collection of apps including EnrichmentMap(Merico et al.), AutoAnnotate(Kucera et al.), WordCloud(Oesper et al.) and clusterMaker2(Morris et al.) used to visualize and analysis enrichment results.
- Install additional cytoscape apps that will be used in this workshop. If using cytoscape 3.6.1 or older the apps need to manually installed through the app manager in Cytoscape or through your web browser. (click on the method to see detailed instructions)
Set up - (part 4)
If you are using Cytoscape 3.7 or higher (Cytoscape 3.7 will be released in August 2018) then apps can be installed directly from R.
#list of cytoscape apps to install
installation_responses <- c()
#list of app to install
cyto_app_toinstall <- c("clustermaker2", "enrichmentmap", "autoannotate", "wordcloud", "stringapp", "aMatReader")
cytoscape_version <- unlist(strsplit( cytoscapeVersionInfo()['cytoscapeVersion'],split = "\\."))
if(length(cytoscape_version) == 3 && as.numeric(cytoscape_version[1]>=3)
&& as.numeric(cytoscape_version[2]>=7)){
for(i in 1:length(cyto_app_toinstall)){
#check to see if the app is installed. Only install it if it hasn't been installed
if(!grep(commandsGET(paste("apps status app=\"", cyto_app_toinstall[i],"\"", sep="")),
pattern = "status: Installed")){
installation_response <-commandsGET(paste("apps install app=\"",
cyto_app_toinstall[i],"\"", sep=""))
installation_responses <- c(installation_responses,installation_response)
} else{
installation_responses <- c(installation_responses,"already installed")
}
}
installation_summary <- data.frame(name = cyto_app_toinstall,
status = installation_responses)
knitr::kable(list(installation_summary),
booktabs = TRUE, caption = 'A Summary of automated app installation'
)
}
Communicating with Cytoscape
Make sure that Cytoscape is open
cytoscapePing ()

Getting started
Confirm that Cytoscape is installed and opened
cytoscapeVersionInfo ()

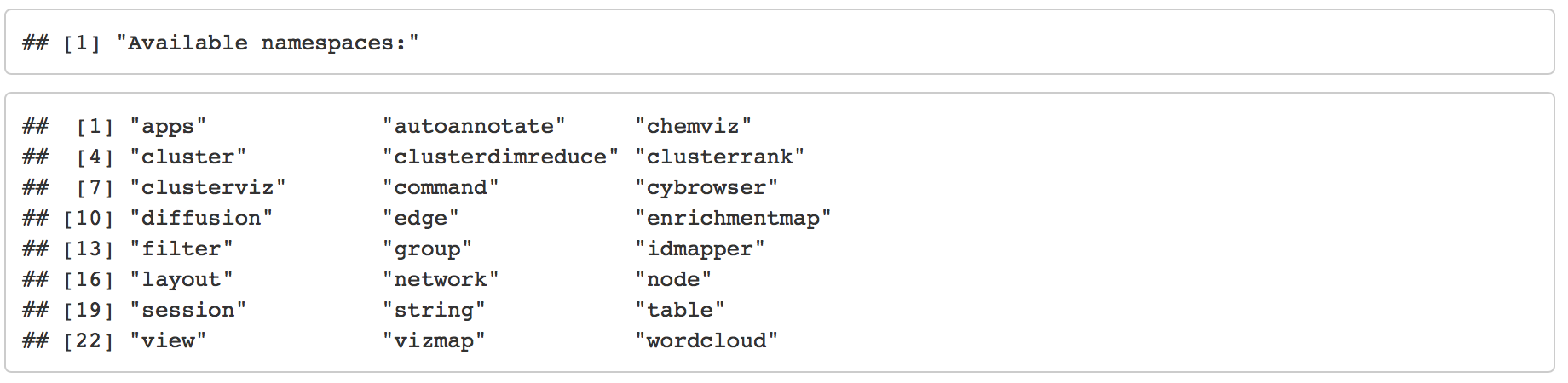
Browse available functions, commands and arguments
Depending on what apps you have installed there is different functionality available.
To see all the functions available in RCy3 package
help(package=RCy3)
cyrestAPI() # CyREST API
commandsAPI() # Commands API
Help on specific cytoscape command
To get information about an individual command from the R environment you can also use the commandsHelp function. Simply specify what command you would like to get information on by adding its name to the command. For example “commandsHelp("help string”)“
commandsHelp("help")
Cytoscape Basics
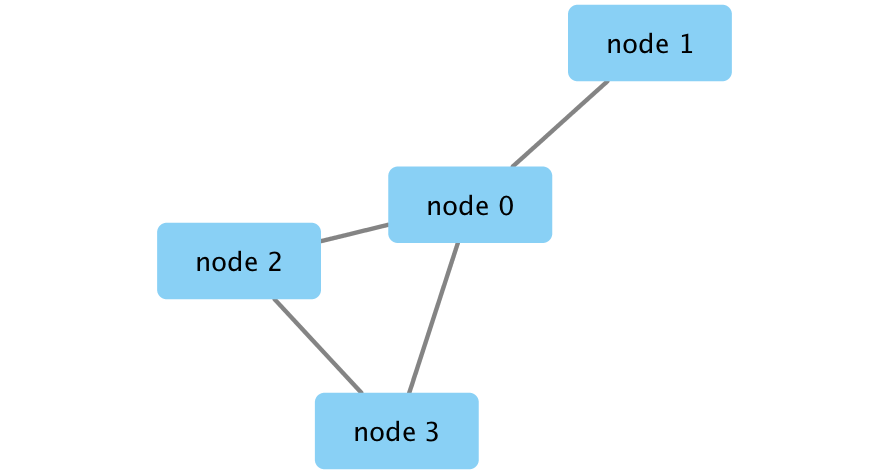
Create a Cytoscape network from some basic R objects
nodes <- data.frame(id=c("node 0","node 1","node 2","node 3"),
group=c("A","A","B","B"), # categorical strings
score=as.integer(c(20,10,15,5)), # integers
stringsAsFactors=FALSE)
edges <- data.frame(source=c("node 0","node 0","node 0","node 2"),
target=c("node 1","node 2","node 3","node 3"),
interaction=c("inhibits","interacts","activates","interacts"), # optional
weight=c(5.1,3.0,5.2,9.9), # numeric
stringsAsFactors=FALSE)
Data frame used to create Network
Nodes table:
| id | group | score |
|---|---|---|
| node 0 | A | 20 |
| node 1 | A | 10 |
| node 2 | B | 15 |
| node 3 | B | 5 |
Edges table:
| source | target | interaction | weight |
|---|---|---|---|
| node 0 | node 1 | inhibits | 5.1 |
| node 0 | node 2 | interacts | 3.0 |
| node 0 | node 3 | activates | 5.2 |
| node 2 | node 3 | interacts | 9.9 |
Create Network
createNetworkFromDataFrames(nodes,edges, title="my first network", collection="DataFrame Example")

Export an image of the network
Remember. All networks we make are created in Cytoscape so get an image of the resulting network and include it in your current analysis if desired.
initial_network_png_file_name <- file.path(getwd(),"230_Isserlin_RCy3_intro", "images","initial_example_network.png")
if(file.exists(initial_network_png_file_name)){
#cytoscape hangs waiting for user response if file already exists. Remove it first
file.remove(initial_network_png_file_name)
}
#export the network
exportImage(initial_network_png_file_name, type = "png")
Initial simple network
Example Data Set
We downloaded gene expression data from the Ovarian Serous Cystadenocarcinoma project of The Cancer Genome Atlas (TCGA)(International Genome et al.), http://cancergenome.nih.gov via the Genomic Data Commons (GDC) portal(Grossman et al.) on 2017-06-14 using TCGABiolinks R package(Colaprico et al.).
- 300 samples available as RNA-seq data
- 79 classified as Immunoreactive, 72 classified as Mesenchymal, 69 classified as Differentiated, and 80 classified as Proliferative samples
- RNA-seq read counts were converted to CPM values and genes with CPM > 1 in at least 50 of the samples are retained for further study
- The data was normalized and differential expression was calculated for each cancer class relative to the rest of the samples.
There are two data files:
- Expression matrix - containing the normalized expression for each gene across all 300 samples.
- Gene ranks - containing the p-values, FDR and foldchange values for the 4 comparisons (mesenchymal vs rest, differential vs rest, proliferative vs rest and immunoreactive vs rest)
RNASeq_expression_matrix <- read.table(
file.path(getwd(),"230_Isserlin_RCy3_intro","data","TCGA_OV_RNAseq_expression.txt"),
header = TRUE, sep = "\t", quote="\"", stringsAsFactors = FALSE)
RNASeq_gene_scores <- read.table(
file.path(getwd(),"230_Isserlin_RCy3_intro","data","TCGA_OV_RNAseq_All_edgeR_scores.txt"),
header = TRUE, sep = "\t", quote="\"", stringsAsFactors = FALSE)
Finding Network Data
How do I represent my data as a network? - Unfortunately, there is not a simple answer.
It depends on your biological question!
Example use cases: - Omics data - I have a ----------- fill in the blank (microarray, RNASeq, Proteomics, ATACseq, MicroRNA, GWAS …) dataset. I have normalized and scored my data. How do I overlay my data on existing interaction data?
- Coexpression data -I have a dataset that represents relationships. How do I represent it as a network.
- Omics data - I have a -----------fill in the blank (microarray, RNASeq, Proteomics, ATACseq, MicroRNA, GWAS …) dataset. I have normalized and scored my data. I have run my data through a functional enrichment tool and now have a set of enriched terms associated with my dataset. How do I represent my functional enrichments as a network?
Use Case 1 - How are my top genes related?
Omics data - I have a -----------fill in the blank (microarray, RNASeq, Proteomics, ATACseq, MicroRNA, GWAS …) dataset. I have normalized and scored my data. How do I overlay my data on existing interaction data?
There are endless amounts of databases storing interaction data.

Cytoscape Apps with network data
Thankfully we don't have to query each independent ally. In addition to many specialized (for example, for specific molecules, interaction type, or species) interaction databases there are also databases that collate these databases to create a broad resource that is easier to use. For example:
- StringApp - is a protein - protein and protein- chemical database that imports data from String(Szklarczyk et al.), [STITCH] into a unified, queriable database.
- PSICQUIC(Aranda et al.) - a REST-ful service that is the responsibility of the database provider to set up and maintain. PSICQUIC is an additional interface that allows users to search all available databases (that support this REST-ful service). The databases are required to represent their interaction data in Proteomic Standards Initiative - molecular interaction (PSI-MI) format. To see a list of all the available data source see here
- nDex(Pratt et al.) - a network data exchange repository.
- GeneMANIA(Mostafavi et al.) - contains multiple networks (shared domains, physical interactions, pathways, predicted, co-expression, genetic interactions and co-localized network). Given a set of genes GeneMANIA selects and weights networks that optimize the connectivity between the query genes. GeneMANIA will also return additional genes that are highly related to your query set.
- PathwayCommons - (access the data through the CyPath2App) is a pathway and interaction data source. Data is collated from a large set of resources (list here ) and stored in the BioPAX(Demir et al.) format. BioPAX is a data standard that allows for detailed representation of pathway mechanistic details as opposed to collapsing it to the set of interactions between molecules. BioPAX pathways from Pathway commons can also be loaded directly into R using the PaxToolsR(Luna et al.) Bioconductor package.
Get a subset of genes of interest from our scored data:
top_mesenchymal_genes <- RNASeq_gene_scores[which(RNASeq_gene_scores$FDR.mesen < 0.05 & RNASeq_gene_scores$logFC.mesen > 2),]
head(top_mesenchymal_genes)
Name geneid logFC.mesen logCPM.mesen LR.mesen PValue.mesen
188 PRG4 10216 2.705305 2.6139056 95.58179 1.42e-22
252 PROK1 84432 2.543381 1.3255202 68.31067 1.40e-16
308 PRRX1 5396 2.077538 4.8570983 123.09925 1.33e-28
434 PTGFR 5737 2.075707 -0.1960881 73.24646 1.14e-17
438 PTGIS 5740 2.094198 5.8279714 165.11038 8.65e-38
1214 BARX1 56033 3.267472 1.7427387 166.30064 4.76e-38
FDR.mesen logFC.diff logCPM.diff LR.diff PValue.diff FDR.diff
188 7.34e-21 -1.2716521 2.6139056 14.107056 1.726950e-04 1.181751e-03
252 4.77e-15 0.7455119 1.3255202 5.105528 2.384972e-02 6.549953e-02
308 1.08e-26 -1.2367108 4.8570983 29.949104 4.440000e-08 1.250000e-06
434 4.30e-16 -0.4233297 -0.1960881 2.318523 1.278413e-01 2.368387e-01
438 1.20e-35 -0.4448761 5.8279714 5.696086 1.700278e-02 5.027140e-02
1214 6.82e-36 -2.4411370 1.7427387 52.224346 4.950000e-13 6.970000e-11
Row.names.y logFC.immuno logCPM.immuno LR.immuno PValue.immuno
188 PRG4|10216 -0.4980017 2.6139056 2.651951 0.103422901
252 PROK1|84432 -1.9692994 1.3255202 27.876348 0.000000129
308 PRRX1|5396 -0.4914091 4.8570983 5.773502 0.016269586
434 PTGFR|5737 -0.6737143 -0.1960881 6.289647 0.012144523
438 PTGIS|5740 -0.6138074 5.8279714 11.708780 0.000622059
1214 BARX1|56033 -0.6063633 1.7427387 4.577141 0.032401228
FDR.immuno logFC.prolif logCPM.prolif LR.prolif PValue.prolif
188 0.185548905 -0.9356510 2.6139056 8.9562066 0.0027652850
252 0.000001650 -1.3195933 1.3255202 13.7675841 0.0002068750
308 0.042062023 -0.3494185 4.8570983 2.9943819 0.0835537930
434 0.033197211 -0.9786631 -0.1960881 12.9112727 0.0003266090
438 0.002779895 -1.0355143 5.8279714 31.9162051 0.0000000161
1214 0.073565295 -0.2199714 1.7427387 0.6315171 0.4267993850
FDR.prolif
188 0.006372597
252 0.000622057
308 0.128528698
434 0.000932503
438 0.000000107
1214 0.510856262
We are going to query the String Database to get all interactions found for our set of top Mesenchymal genes.
commandsHelp("help string")

commandsHelp("help string protein query")

Query String
mesen_string_interaction_cmd <- paste('string protein query taxonID=9606 limit=150 cutoff=0.9 query="',paste(top_mesenchymal_genes$Name, collapse=","),'"',sep="")
commandsGET(mesen_string_interaction_cmd)

Get a screenshot of the initial network
initial_string_network_png_file_name <- file.path(getwd(),"230_Isserlin_RCy3_intro", "images", "initial_string_network.png")
if(file.exists(initial_string_network_png_file_name)){
#cytoscape hangs waiting for user response if file already exists. Remove it first
response <- file.remove(initial_string_network_png_file_name)
}
response <- exportImage(initial_string_network_png_file_name, type = "png")
Initial String Network - top Mesenchymal genes interactions
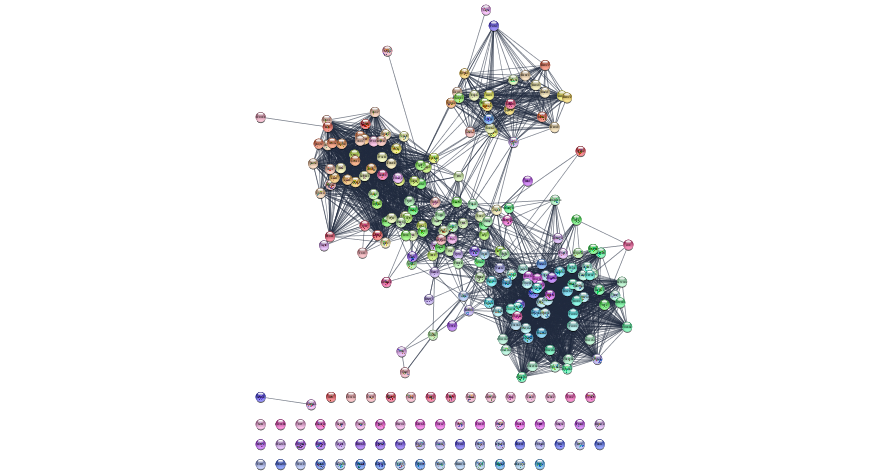
Layouts
Layout the network
layoutNetwork('force-directed')
Check what other layout algorithms are available to try out
getLayoutNames()

Layouts - cont'd
Get the parameters for a specific layout
getLayoutPropertyNames(layout.name='force-directed')

Layouts - cont'd
Re-layout the network using the force directed layout but specify some of the parameters
layoutNetwork('force-directed defaultSpringCoefficient=0.0000008 defaultSpringLength=70')
Get a screenshot of the re-laid out network
relayout_string_network_png_file_name <- file.path(getwd(),"230_Isserlin_RCy3_intro",
"images","relayout_string_network.png")
if(file.exists(relayout_string_network_png_file_name)){
#cytoscape hangs waiting for user response if file already exists. Remove it first
response<- file.remove(relayout_string_network_png_file_name)
}
response <- exportImage(relayout_string_network_png_file_name, type = "png")
String network with new layout

Overlay our expression data on the String network.
To do this we will be using the loadTableData function from RCy3. It is important to make sure that that your identifiers types match up. You can check what is used by String by pulling in the column names of the node attribute table.
getTableColumnNames('node')

Overlay our expression data on the String network - cont'd
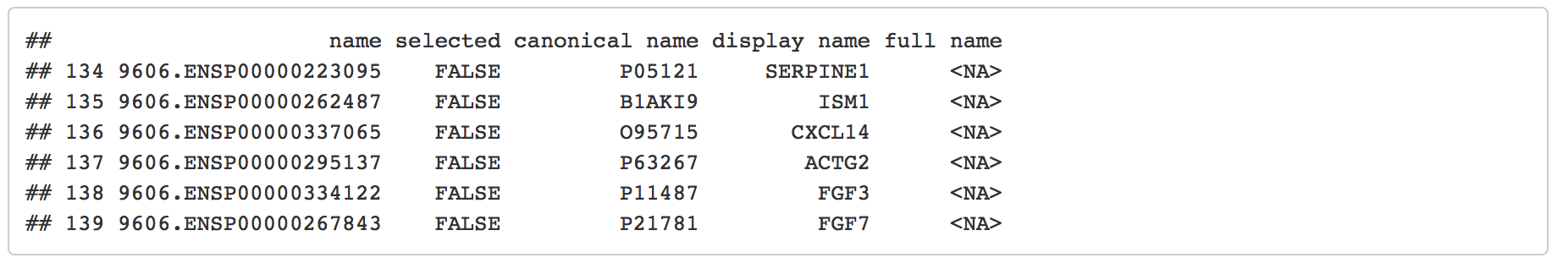
If you are unsure of what each column is and want to further verify the column to use you can also pull in the entire node attribute table.
node_attribute_table_topmesen <- getTableColumns(table="node")
head(node_attribute_table_topmesen[,3:7])
Overlay our expression data on the String network - cont'd
The column “display name” contains HGNC gene names which are also found in our Ovarian Cancer dataset.
To import our expression data we will match our dataset to the “display name” node attribute.
?loadTableData
loadTableData(RNASeq_gene_scores,table.key.column = "display name",
data.key.column = "Name") #default data.frame key is row.names

Visual Style
Modify the visual style Create your own visual style to visualize your expression data on the String network.
Start with a default style
style.name = "MesenchymalStyle"
defaults.list <- list(NODE_SHAPE="ellipse",
NODE_SIZE=60,
NODE_FILL_COLOR="#AAAAAA",
EDGE_TRANSPARENCY=120)
# p for passthrough; nothing else needed
node.label.map <- mapVisualProperty('node label','display name','p')
createVisualStyle(style.name, defaults.list, list(node.label.map))
setVisualStyle(style.name=style.name)

Visual Style - cont'd
Update your created style with a mapping for the Mesenchymal logFC expression. The first step is to grab the column data from Cytoscape (we can reuse the node_attribute table concept from above but we have to call the function again as we have since added our expression data) and pull out the min and max to define our data mapping range of values.
Note: you could define the min and max based on the entire dataset or just the subset that is represented in Cytoscape currently. The two methods will give you different results. If you intend on comparing different networks created with the same dataset then it is best to calculate the min and max from the entire dataset as opposed to a subset.
min.mesen.logfc = min(RNASeq_gene_scores$logFC.mesen,na.rm=TRUE)
max.mesen.logfc = max(RNASeq_gene_scores$logFC.mesen,na.rm=TRUE)
data.values = c(min.mesen.logfc,0,max.mesen.logfc)
Visual Style - cont'd
Next, we use the RColorBrewer package to help us pick good colors to pair with our data values.
library(RColorBrewer)
display.brewer.all(length(data.values), colorblindFriendly=TRUE, type="div")
logFC to node color
node.colors <- c(rev(brewer.pal(length(data.values), "RdBu")))
Map the colors to our data value and update our visual style.
setNodeColorMapping("logFC.mesen", data.values, node.colors, style.name=style.name)
Visual Style - cont'd
Remember, String includes your query proteins as well as other proteins that associate with your query proteins (including the strongest connection first). Not all of the proteins in this network are your top hits. How can we visualize which proteins are our top Mesenchymal hits?
Add a different border color or change the node shape for our top hits.
getNodeShapes()
#select the Nodes of interest
#selectNode(nodes = top_mesenchymal_genes$Name, by.col="display name")
setNodeShapeBypass(node.names = top_mesenchymal_genes$Name, new.shapes = "TRIANGLE")

Visual Style - cont'd
Change the size of the node to be correlated with the Mesenchymal p-value.
setNodeSizeMapping(table.column = 'LR.mesen',
table.column.values = c(min(RNASeq_gene_scores$LR.mesen),
mean(RNASeq_gene_scores$LR.mesen),
max(RNASeq_gene_scores$LR.mesen)),
sizes = c(30, 60, 150),mapping.type = "c", style.name = style.name)
Visual Style - cont'd
Get a screenshot of the resulting network
mesen_string_network_png_file_name <- file.path(getwd(),"230_Isserlin_RCy3_intro", "images","mesen_string_network.png")
if(file.exists(mesen_string_network_png_file_name)){
#cytoscape hangs waiting for user response if file already exists. Remove it first
response<- file.remove(mesen_string_network_png_file_name)
}
response <- exportImage(mesen_string_network_png_file_name, type = "png")
Formatted String network
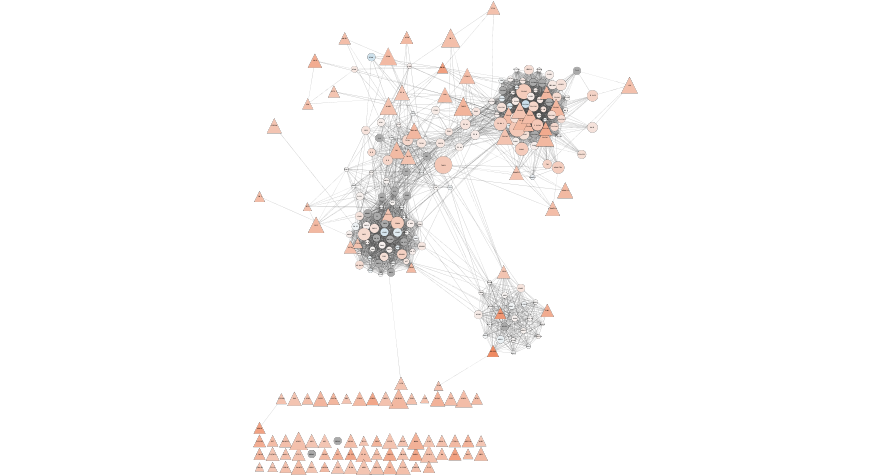
Use Case 2 - Which genes have similar expression.
Instead of querying existing resources look for correlations in your own dataset to find out which genes have similar expression. There are many tools that can analyze your data for correlation. A popular tool is Weighted Gene Correlation Network Analysis (WGCNA)(Langfelder et al.) which takes expression data and calculates functional modules. As a simple example we can transform our expression dataset into a correlation matrix.
Using the Cytoscape App, aMatReader(Settle et al.), we transform our adjacency matrix into an interaction network. First we filter the correlation matrix to contain only the strongest connections (for example, only correlations greater than 0.9).
RNASeq_expression <- RNASeq_expression_matrix[,3:ncol(RNASeq_expression_matrix)]
rownames(RNASeq_expression) <- RNASeq_expression_matrix$Name
RNAseq_correlation_matrix <- cor(t(RNASeq_expression), method="pearson")
#set the diagonal of matrix to zero - eliminate self-correlation
RNAseq_correlation_matrix[
row(RNAseq_correlation_matrix) == col(RNAseq_correlation_matrix) ] <- 0
# set all correlations that are less than 0.9 to zero
RNAseq_correlation_matrix[which(RNAseq_correlation_matrix<0.90)] <- 0
#get rid of rows and columns that have no correlations with the above thresholds
RNAseq_correlation_matrix <- RNAseq_correlation_matrix[which(rowSums(RNAseq_correlation_matrix) != 0),
which(colSums(RNAseq_correlation_matrix) !=0)]
#write out the correlation file
correlation_filename <- file.path(getwd(), "230_Isserlin_RCy3_intro", "data", "TCGA_OV_RNAseq_expression_correlation_matrix.txt")
write.table( RNAseq_correlation_matrix, file = correlation_filename, col.names = TRUE, row.names = FALSE, sep = "\t", quote=FALSE)
Use the CyRest call to access the aMatReader functionality.
amat_url <- "aMatReader/v1/import"
amat_params = list(files = list(correlation_filename),
delimiter = "TAB",
undirected = FALSE,
ignoreZeros = TRUE,
interactionName = "correlated with",
rowNames = FALSE
)
response <- cyrestPOST(operation = amat_url, body = amat_params,
base.url = "http://localhost:1234")
current_network_id <- response$data["suid"]
Modify resulting network
Relayout
#relayout network
layoutNetwork('cose',
network = as.numeric(current_network_id))
Rename
renameNetwork(title = "Coexpression_network_pear0_9",
network = as.numeric(current_network_id))
Modify resulting network - cont'd
Modify the visualization to see where each genes is predominantly expressed. Look at the 4 different p-values associated with each gene and color the nodes with the type associated with the lowest FDR.
Load in the scoring data. Specify the cancer type where the genes has the lowest FDR value.
nodes_in_network <- rownames(RNAseq_correlation_matrix)
#add an additional column to the gene scores table to indicate in which samples
# the gene is significant
node_class <- vector(length = length(nodes_in_network),mode = "character")
for(i in 1:length(nodes_in_network)){
current_row <- which(RNASeq_gene_scores$Name == nodes_in_network[i])
min_pvalue <- min(RNASeq_gene_scores[current_row,
grep(colnames(RNASeq_gene_scores), pattern = "FDR")])
if(RNASeq_gene_scores$FDR.mesen[current_row] <=min_pvalue){
node_class[i] <- paste(node_class[i],"mesen",sep = " ")
}
if(RNASeq_gene_scores$FDR.diff[current_row] <=min_pvalue){
node_class[i] <- paste(node_class[i],"diff",sep = " ")
}
if(RNASeq_gene_scores$FDR.prolif[current_row] <=min_pvalue){
node_class[i] <- paste(node_class[i],"prolif",sep = " ")
}
if(RNASeq_gene_scores$FDR.immuno[current_row] <=min_pvalue){
node_class[i] <- paste(node_class[i],"immuno",sep = " ")
}
}
node_class <- trimws(node_class)
node_class_df <-data.frame(name=nodes_in_network, node_class,stringsAsFactors = FALSE)
head(node_class_df)
name node_class
1 ABCA6 mesen
2 ABCA8 mesen
3 ABI3 immuno
4 ACAN prolif
5 ACAP1 immuno
6 ADAM12 mesen
Modify resulting network - cont'd
Map the new node attribute and the all the gene scores to the network.
#default data.frame key is row.names
loadTableData(RNASeq_gene_scores,table.key.column = "name",data.key.column = "Name")
#default data.frame key is row.names
loadTableData(node_class_df,table.key.column = "name",data.key.column = "name")
Modify resulting network - cont'd
Create a color mapping for the different cancer types.
#create a new mapping with the different types
unique_types <- sort(unique(node_class))
coul = brewer.pal(4, "Set1")
# I can add more tones to this palette :
coul = colorRampPalette(coul)(length(unique_types))
setNodeColorMapping(table.column = "node_class",table.column.values = unique_types,
colors = coul,mapping.type = "d")
Modify resulting network - cont'd
correlation_network_png_file_name <- file.path(getwd(),"230_Isserlin_RCy3_intro", "images","correlation_network.png")
if(file.exists(correlation_network_png_file_name)){
#cytoscape hangs waiting for user response if file already exists. Remove it first
file.remove(correlation_network_png_file_name)
}
#export the network
exportImage(correlation_network_png_file_name, type = "png")
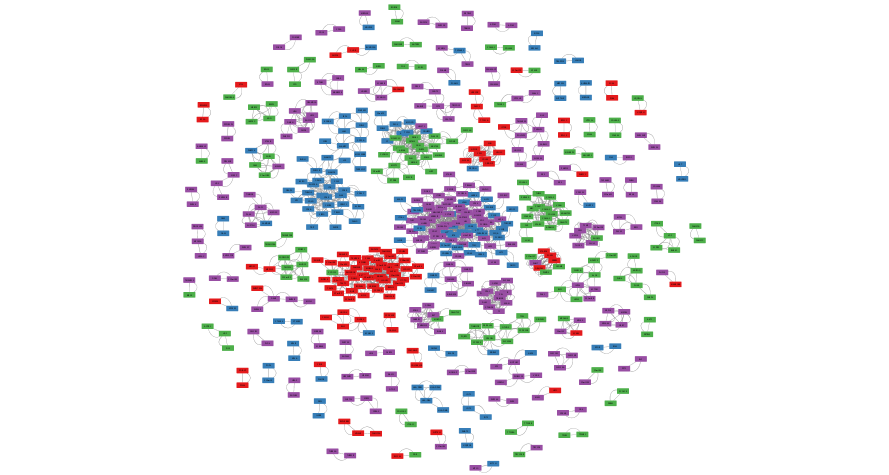
Cluster the Network
#make sure it is set to the right network
setCurrentNetwork(network = getNetworkName(suid=as.numeric(current_network_id)))
#cluster the network
clustermaker_url <- paste("cluster mcl network=SUID:",current_network_id, sep="")
commandsGET(clustermaker_url)
#get the clustering results
default_node_table <- getTableColumns(table= "node",network = as.numeric(current_network_id))
head(default_node_table)
Enrichment Analysis
Perform pathway Enrichment on one of the clusters using g:Profiler(Reimand et al.). g:Profiler is an online functional enrichment web service that will take your gene list and return the set of enriched pathways. For automated analysis g:Profiler has created an R library to interact with it directly from R instead of using the web page.
Create a function to call g:Profiler and convert the returned results into a generic enrichment map input file.
tryCatch(expr = { library("gProfileR")},
error = function(e) { install.packages("gProfileR")}, finally = library("gProfileR"))
#function to run gprofiler using the gprofiler library
#
# The function takes the returned gprofiler results and formats it to the generic EM input file
#
# function returns a data frame in the generic EM file format.
runGprofiler <- function(genes,current_organism = "hsapiens",
significant_only = F, set_size_max = 200,
set_size_min = 3, filter_gs_size_min = 5 , exclude_iea = F){
gprofiler_results <- gprofiler(genes ,
significant=significant_only,ordered_query = F,
exclude_iea=exclude_iea,max_set_size = set_size_max,
min_set_size = set_size_min,
correction_method = "fdr",
organism = current_organism,
src_filter = c("GO:BP","REAC"))
#filter results
gprofiler_results <- gprofiler_results[which(gprofiler_results[,'term.size'] >= 3
& gprofiler_results[,'overlap.size'] >= filter_gs_size_min ),]
# gProfileR returns corrected p-values only. Set p-value to corrected p-value
if(dim(gprofiler_results)[1] > 0){
em_results <- cbind(gprofiler_results[,
c("term.id","term.name","p.value","p.value")], 1,
gprofiler_results[,"intersection"])
colnames(em_results) <- c("Name","Description", "pvalue","qvalue","phenotype","genes")
return(em_results)
} else {
return("no gprofiler results for supplied query")
}
}
Enrichment Analysis - cont'd
Run g:Profiler. g:Profiler will return a set of pathways and functions that are found to be enriched in our query set of genes.
current_cluster <- "1"
#select all the nodes in cluster 1
selectednodes <- selectNodes(current_cluster, by.col="__mclCluster")
#create a subnetwork with cluster 1
subnetwork_suid <- createSubnetwork(nodes="selected")
renameNetwork("Cluster1_Subnetwork", network=as.numeric(subnetwork_suid))
subnetwork_node_table <- getTableColumns(table= "node",network = as.numeric(subnetwork_suid))
em_results <- runGprofiler(subnetwork_node_table$name)
#write out the g:Profiler results
em_results_filename <-file.path(getwd(),
"230_Isserlin_RCy3_intro", "data",paste("gprofiler_cluster",current_cluster,"enr_results.txt",sep="_"))
write.table(em_results,em_results_filename,col.name=TRUE,sep="\t",row.names=FALSE,quote=FALSE)
head(em_results)
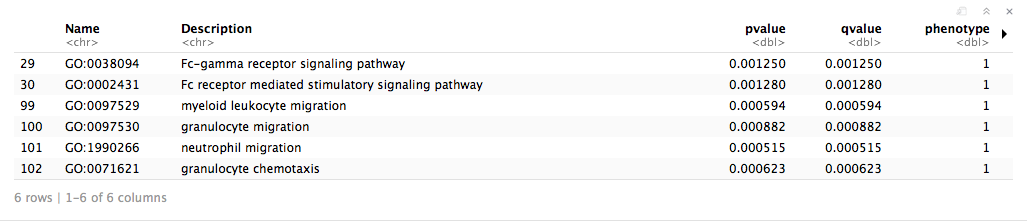
Enrichment Analysis - cont'd
Create an enrichment map with the returned g:Profiler results. An enrichment map is a different sort of network. Instead of nodes representing genes, nodes represent pathways or functions. Edges between these pathways or functions represent shared genes or pathway crosstalk. An enrichment map is a way to visualize your enrichment results to help reduce redundancy and uncover main themes. Pathways can also be explored in detail using the features available through the App in Cytoscape.
em_command = paste('enrichmentmap build analysisType="generic" ',
'pvalue=',"0.05", 'qvalue=',"0.05",
'similaritycutoff=',"0.25",
'coeffecients=',"JACCARD",
'enrichmentsDataset1=',em_results_filename ,
sep=" ")
#enrichment map command will return the suid of newly created network.
em_network_suid <- commandsRun(em_command)
renameNetwork("Cluster1_enrichmentmap", network=as.numeric(em_network_suid))
Enrichment Analysis - cont'd
Export image of resulting Enrichment map.
cluster1em_png_file_name <- file.path(getwd(),"230_Isserlin_RCy3_intro", "images","cluster1em.png")
if(file.exists(cluster1em_png_file_name)){
#cytoscape hangs waiting for user response if file already exists. Remove it first
file.remove(cluster1em_png_file_name)
}
#export the network
exportImage(cluster1em_png_file_name, type = "png")
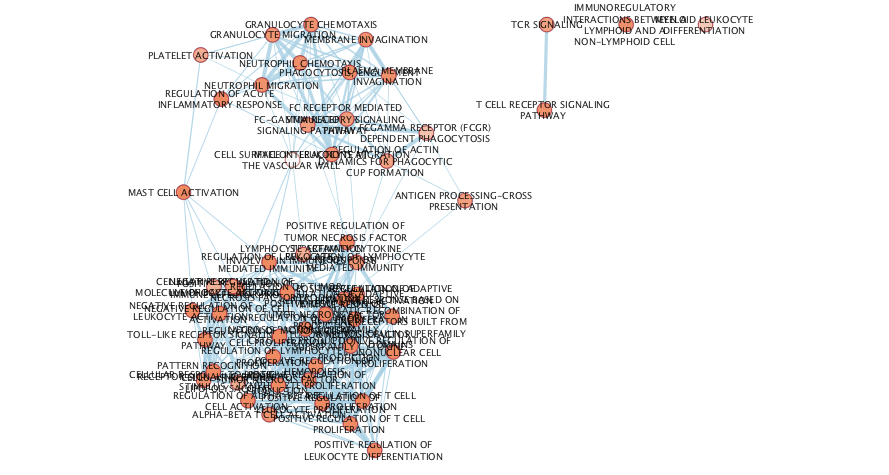
Enrichment Analysis - cont'd
Annotate the Enrichment map to get the general themes that are found in the enrichment results of cluster 1
#get the column from the nodetable and node table
nodetable_colnames <- getTableColumnNames(table="node", network = as.numeric(em_network_suid))
descr_attrib <- nodetable_colnames[grep(nodetable_colnames, pattern = "_GS_DESCR")]
#Autoannotate the network
autoannotate_url <- paste("autoannotate annotate-clusterBoosted labelColumn=", descr_attrib," maxWords=3 ", sep="")
current_name <-commandsGET(autoannotate_url)
Enrichment Analysis - cont'd
Export image of resulting Annotated Enrichment map.
cluster1em_annot_png_file_name <- file.path(getwd(),"230_Isserlin_RCy3_intro", "images","cluster1em_annot.png")
if(file.exists(cluster1em_annot_png_file_name)){
#cytoscape hangs waiting for user response if file already exists. Remove it first
file.remove(cluster1em_annot_png_file_name)
}
#export the network
exportImage(cluster1em_annot_png_file_name, type = "png")
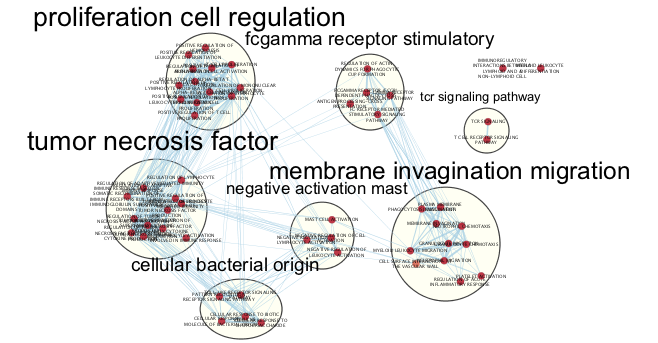
Enrichment Analysis - cont'd
Dense networks small or large never look like network figures we so often see in journals. A lot of manual tweaking, reorganization and optimization is involved in getting that perfect figure ready network. The above network is what the network starts as. The below figure is what it can look like after a few minutes of manual reorganiazation. (individual clusters were selected from the auto annotate panel and separated from other clusters)
Use Case 3 - Functional Enrichment of Omics set.
Reducing our large OMICs expression set to a simple list of genes eliminates the majority of the signals present in the data. Thresholding will only highlight the strong signals neglecting the often more interesting subtle signals. In order to capitalize on the wealth of data present in the data we need to perform pathway enrichment analysis on the entire expression set. There are many tools in R or as standalone apps that perform this type of analysis.
To demonstrate how you can use Cytoscape and RCy3 in your enrichment analysis pipeline we will use EnrichmentBrowser package (as demonstrated in detail in the workshop Functional enrichment analysis of high-throughput omics data) to perform pathway analysis.
if(!"EnrichmentBrowser" %in% installed.packages()){
install.packages("BiocManager")
BiocManager::install("EnrichmentBrowser")
}
suppressPackageStartupMessages(library(EnrichmentBrowser))
Geneset Files
Download the latest pathway definition file from the Baderlab download site. Baderlab genesets are updated on a monthly basis. Detailed information about the sources can be found here.
Only Human, Mouse and Rat gene set files are currently available on the Baderlab downloads site. If you are working with a species other than human (and it is either rat or mouse) change the gmt_url below to correct species.
Geneset Files - cont'd
tryCatch(expr = { suppressPackageStartupMessages(library("RCurl"))},
error = function(e) { install.packages("RCurl")},
finally = library("RCurl"))
gmt_url = "http://download.baderlab.org/EM_Genesets/current_release/Human/symbol/"
#list all the files on the server
filenames = getURL(gmt_url)
tc = textConnection(filenames)
contents = suppressWarnings(readLines(tc))
close(tc)
#get the gmt that has all the pathways and does not include terms inferred from electronic annotations(IEA)
#start with gmt file that has pathways only
rx = gregexpr("(?<=<a href=\")(.*.GOBP_AllPathways_no_GO_iea.*.)(.gmt)(?=\">)",
contents, perl = TRUE)
gmt_file = unlist(regmatches(contents, rx))
dest_gmt_file <- file.path(getwd(), "230_Isserlin_RCy3_intro", "data", gmt_file)
download.file(
paste(gmt_url,gmt_file,sep=""),
destfile=dest_gmt_file
)
Geneset Files - cont'd
Load in the gmt file
library(EnrichmentBrowser)
baderlab.gs <- getGenesets(dest_gmt_file)
baderlab.gs[1:2]
$`THIO-MOLYBDENUM COFACTOR BIOSYNTHESIS%HUMANCYC%PWY-5963`
[1] "MOCOS"
$`PROLINE BIOSYNTHESIS I%HUMANCYC%PROSYN-PWY`
[1] "PYCR2" "ALDH18A1" "PYCR1" "PYCRL"
EnrichmentBrowser
Create the dataset required by EnrichmentBrowser tools
#create the expression file - A tab separated text file containing expression values. Columns = samples/subjects; rows = features/probes/genes; NO headers, row or column names.
expr <- RNASeq_expression
sumexpr_filename <- file.path(getwd(), "230_Isserlin_RCy3_intro","data","SummarizeExperiment_expression.txt")
write.table( expr , file = sumexpr_filename , col.names = FALSE, row.names = FALSE, sep = "\t", quote=FALSE)
rowData <- RNASeq_gene_scores[,grep(colnames(RNASeq_gene_scores), pattern="mesen")]
rowData <- cbind(RNASeq_gene_scores$Name,rowData)
colnames(rowData)[2] <- "FC"
colnames(rowData)[6] <- "ADJ.PVAL"
sumexpr_rdat_filename <- file.path(getwd(), "230_Isserlin_RCy3_intro","data","SummarizeExperiment_rdat.txt")
write.table( rowData[,1] , file = sumexpr_rdat_filename , col.names = FALSE, row.names = FALSE, sep = "\t", quote=FALSE)
#load in the data classification data
# A tab separated text file containing annotation information for the samples in either *two or three* columns. NO headers, row or column names. The number of rows/samples in this file should match the number of columns/samples of the expression matrix. The 1st column is reserved for the sample IDs; The 2nd column is reserved for a *BINARY* group assignment. Use '0' and '1' for unaffected (controls) and affected (cases) sample class
classDefinitions_RNASeq <- read.table(
file.path(getwd(), "230_Isserlin_RCy3_intro","data","RNASeq_classdefinitions.txt"), header = TRUE, sep = "\t", quote="\"", stringsAsFactors = FALSE)
colData <- data.frame(Sample = colnames(RNASeq_expression),
GROUP = classDefinitions_RNASeq$SUBTYPE,
stringsAsFactors = FALSE)
rownames(colData) <- colnames(RNASeq_expression)
colData$GROUP[which(colData$GROUP != "Mesenchymal")] <- 0
colData$GROUP[which(colData$GROUP == "Mesenchymal")] <- 1
sumexpr_cdat_filename <- file.path(getwd(), "230_Isserlin_RCy3_intro","data","SummarizeExperiment_cdat.txt")
write.table( colData , file = sumexpr_cdat_filename , col.names = FALSE, row.names = FALSE, sep = "\t", quote=FALSE)
#create the Summarize Experiment object
se_OV <- EnrichmentBrowser::readSE(assay.file = sumexpr_filename , cdat.file = sumexpr_cdat_filename, rdat.file = sumexpr_rdat_filename)
EnrichmentBrowser - cont'd
Put our precomputed p-values and fold change values into the Summarized Experiment object so we can use our rankings for the analysis
#set the Summarized Experiment to our computed p-values and FC
rowData(se_OV) <- rowData
EnrichmentBrowser - cont'd
Run basic Over representation analysis (ORA) using our ranked genes and our gene set file downloaded from the Baderlab genesets.
ora.all <- sbea(method="ora", se=se_OV, gs=baderlab.gs, perm=0, alpha=0.05)
gsRanking(ora.all)

EnrichmentBrowser - cont'd
Take the enrichment results and create a generic enrichment map input file so we can create an Enrichment map. Description of format of the generic input file can be found here and example generic enrichment map files can be found here
#manually adjust p-values
ora.all$res.tbl <- cbind(ora.all$res.tbl, p.adjust(ora.all$res.tbl$P.VALUE, "BH"))
colnames(ora.all$res.tbl)[ncol(ora.all$res.tbl)] <- "Q.VALUE"
#create a generic enrichment map file
em_results_mesen <- data.frame(name = ora.all$res.tbl$GENE.SET,descr = ora.all$res.tbl$GENE.SET,
pvalue=ora.all$res.tbl$P.VALUE, qvalue=ora.all$res.tbl$Q.VALUE, stringsAsFactors = FALSE)
#write out the ora results
em_results_mesen_filename <-file.path(getwd(),
"230_Isserlin_RCy3_intro","data","mesen_ora_enr_results.txt")
write.table(em_results_mesen,em_results_mesen_filename,col.name=TRUE,sep="\t",row.names=FALSE,quote=FALSE)
Enrichment Map
Create an enrichment map with the returned ORA results.
em_command = paste('enrichmentmap build analysisType="generic" ', "gmtFile=", dest_gmt_file,
'pvalue=',"0.05", 'qvalue=',"0.05",
'similaritycutoff=',"0.25",
'coeffecients=',"JACCARD",
'enrichmentsDataset1=',em_results_mesen_filename ,
sep=" ")
#enrichment map command will return the suid of newly created network.
em_mesen_network_suid <- commandsRun(em_command)
renameNetwork("Mesenchymal_ORA_enrichmentmap", network=as.numeric(em_mesen_network_suid))
Enrichment Map - cont'd
Export image of resulting Enrichment map.
mesenem_png_file_name <- file.path(getwd(),"230_Isserlin_RCy3_intro", "images","mesenem.png")
if(file.exists(mesenem_png_file_name)){
#cytoscape hangs waiting for user response if file already exists. Remove it first
file.remove(mesenem_png_file_name)
}
#export the network
exportImage(mesenem_png_file_name, type = "png")
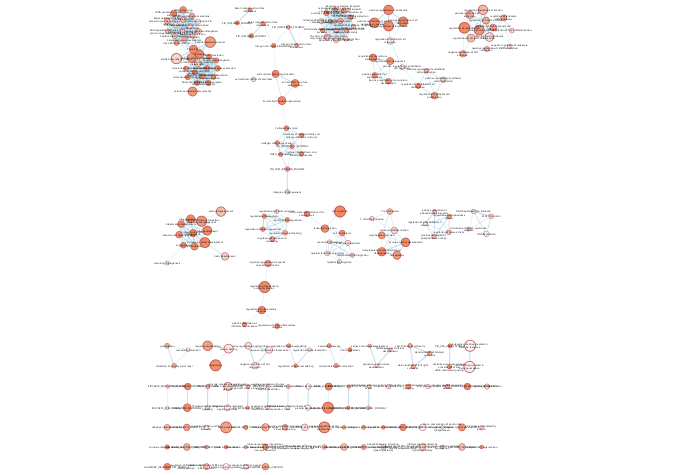
Enrichment Map - cont'd
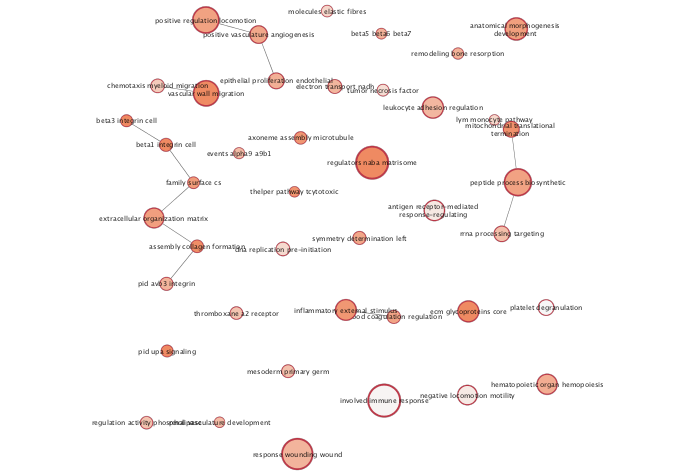
Annotate the Enrichment map to get the general themes that are found in the enrichment results. Often for very busy networks annotating the networks doesn't help to reduce the complexity but instead adds to it. To get rid of some of the pathway redundancy and density in the network create a summary of the underlying network. The summary network collapses each cluster to a summary node. Each summary node is annotated with a word tag (the top 3 words associated with the nodes of the cluster) that is computed using the Wordcloud app.
#get the column from the nodetable and node table
nodetable_colnames <- getTableColumnNames(table="node", network = as.numeric(em_mesen_network_suid))
descr_attrib <- nodetable_colnames[grep(nodetable_colnames, pattern = "_GS_DESCR")]
#Autoannotate the network
autoannotate_url <- paste("autoannotate annotate-clusterBoosted labelColumn=", descr_attrib," maxWords=3 ", sep="")
current_name <-commandsGET(autoannotate_url)
#create a summary network
commandsGET("autoannotate summary network='current'")
#change the network name
summary_network_suid <- getNetworkSuid()
renameNetwork(title = "Mesen_ORA_summary_netowrk",
network = as.numeric(summary_network_suid))
#get the summary node names
summary_nodes <- getTableColumns(table="node",columns=c("name"))
#clear bypass style the summary network has
clearNodePropertyBypass(node.names = summary_nodes$name,visual.property = "NODE_SIZE")
#relayout network
layoutNetwork('cose',
network = as.numeric(summary_network_suid))
Enrichment Map - cont'd
Export image of resulting Summarized Annotated Enrichment map.
mesenem_summary_png_file_name <- file.path(getwd(),"230_Isserlin_RCy3_intro","images","mesenem_summary_network.png")
if(file.exists(mesenem_summary_png_file_name)){
#cytoscape hangs waiting for user response if file already exists. Remove it first
file.remove(mesenem_summary_png_file_name)
}
#export the network
exportImage(mesenem_summary_png_file_name, type = "png")
References
Aranda, Bruno, Hagen Blankenburg, Samuel Kerrien, Fiona SL Brinkman, Arnaud Ceol, Emilie Chautard, Jose M Dana, et al. 2011. “PSICQUIC and Psiscore: Accessing and Scoring Molecular Interactions.” Nature Methods 8 (7). Nature Publishing Group: 528.
Colaprico, A., T. C. Silva, C. Olsen, L. Garofano, C. Cava, D. Garolini, T. S. Sabedot, et al. 2016. “TCGAbiolinks: An R/Bioconductor Package for Integrative Analysis of Tcga Data.” Journal Article. Nucleic Acids Res 44 (8): e71. doi:10.1093/nar/gkv1507.
Demir, Emek, Michael P Cary, Suzanne Paley, Ken Fukuda, Christian Lemer, Imre Vastrik, Guanming Wu, et al. 2010. “The Biopax Community Standard for Pathway Data Sharing.” Nature Biotechnology 28 (9). Nature Publishing Group: 935.
Grossman, R. L., A. P. Heath, V. Ferretti, H. E. Varmus, D. R. Lowy, W. A. Kibbe, and L. M. Staudt. 2016. “Toward a Shared Vision for Cancer Genomic Data.” Journal Article. N Engl J Med 375 (12): 1109–12. doi:10.1056/NEJMp1607591.
International Cancer Genome, Consortium, T. J. Hudson, W. Anderson, A. Artez, A. D. Barker, C. Bell, R. R. Bernabe, et al. 2010. “International Network of Cancer Genome Projects.” Journal Article. Nature 464 (7291): 993–8. doi:10.1038/nature08987.
Kucera, Mike, Ruth Isserlin, Arkady Arkhangorodsky, and Gary D Bader. 2016. “AutoAnnotate: A Cytoscape App for Summarizing Networks with Semantic Annotations.” F1000Research 5. Faculty of 1000 Ltd.
Langfelder, Peter, and Steve Horvath. 2008. “WGCNA: An R Package for Weighted Correlation Network Analysis.” BMC Bioinformatics 9 (1). BioMed Central: 559.
Li, B., and C. N. Dewey. 2011. “RSEM: Accurate Transcript Quantification from Rna-Seq Data with or Without a Reference Genome.” Journal Article. BMC Bioinformatics 12: 323. doi:10.1186/1471-2105-12-323.
Luna, Augustin, Özgün Babur, Bülent Arman Aksoy, Emek Demir, and Chris Sander. 2015. “PaxtoolsR: Pathway Analysis in R Using Pathway Commons.” Bioinformatics 32 (8). Oxford University Press: 1262–4.
Merico, Daniele, Ruth Isserlin, Oliver Stueker, Andrew Emili, and Gary D Bader. 2010. “Enrichment Map: A Network-Based Method for Gene-Set Enrichment Visualization and Interpretation.” PloS One 5 (11). Public Library of Science: e13984.
Morris, John H, Leonard Apeltsin, Aaron M Newman, Jan Baumbach, Tobias Wittkop, Gang Su, Gary D Bader, and Thomas E Ferrin. 2011. “ClusterMaker: A Multi-Algorithm Clustering Plugin for Cytoscape.” BMC Bioinformatics 12 (1). BioMed Central: 436.
Mostafavi, Sara, Debajyoti Ray, David Warde-Farley, Chris Grouios, and Quaid Morris. 2008. “GeneMANIA: A Real-Time Multiple Association Network Integration Algorithm for Predicting Gene Function.” Genome Biology 9 (1). BioMed Central: S4.
Oesper, Layla, Daniele Merico, Ruth Isserlin, and Gary D Bader. 2011. “WordCloud: A Cytoscape Plugin to Create a Visual Semantic Summary of Networks.” Source Code for Biology and Medicine 6 (1). BioMed Central: 7.
Ono, Keiichiro, Tanja Muetze, Georgi Kolishovski, Paul Shannon, and Barry Demchak. 2015. “CyREST: Turbocharging Cytoscape Access for External Tools via a Restful Api.” F1000Research 4. Faculty of 1000 Ltd.
Pratt, Dexter, Jing Chen, Rudolf Pillich, Vladimir Rynkov, Aaron Gary, Barry Demchak, and Trey Ideker. 2017. “NDEx 2.0: A Clearinghouse for Research on Cancer Pathways.” Cancer Research 77 (21). AACR: e58–e61.
Reimand, Jüri, Tambet Arak, Priit Adler, Liis Kolberg, Sulev Reisberg, Hedi Peterson, and Jaak Vilo. 2016. “G: Profiler—a Web Server for Functional Interpretation of Gene Lists (2016 Update).” Nucleic Acids Research 44 (W1). Oxford University Press: W83–W89.
Robinson, M. D., D. J. McCarthy, and G. K. Smyth. 2010. “EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data.” Journal Article. Bioinformatics 26 (1): 139–40. doi:10.1093/bioinformatics/btp616.
Shannon, Paul, Andrew Markiel, Owen Ozier, Nitin S Baliga, Jonathan T Wang, Daniel Ramage, Nada Amin, Benno Schwikowski, and Trey Ideker. 2003. “Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks.” Genome Research 13 (11). Cold Spring Harbor Lab: 2498–2504.
Szklarczyk, Damian, John H Morris, Helen Cook, Michael Kuhn, Stefan Wyder, Milan Simonovic, Alberto Santos, et al. 2016. “The String Database in 2017: Quality-Controlled Protein–protein Association Networks, Made Broadly Accessible.” Nucleic Acids Research. Oxford University Press, gkw937.
Verhaak, R. G., P. Tamayo, J. Y. Yang, D. Hubbard, H. Zhang, C. J. Creighton, S. Fereday, et al. 2013. “Prognostically Relevant Gene Signatures of High-Grade Serous Ovarian Carcinoma.” Journal Article. J Clin Invest 123 (1): 517–25. doi:10.1172/JCI65833.
Wang, K., D. Singh, Z. Zeng, S. J. Coleman, Y. Huang, G. L. Savich, X. He, et al. 2010. “MapSplice: Accurate Mapping of Rna-Seq Reads for Splice Junction Discovery.” Journal Article. Nucleic Acids Res 38 (18): e178. doi:10.1093/nar/gkq622.