Cytoscape Futures
John "Scooter" Morris
EBI Single Cell Workshop
26-31 April 2021
Cytoscape 3.9
- Major new features:
- Table styles -- can set up mappings for table columns
- Improved performance for large networks
- Support for edge Z order
- Haystack edges
- Copy/paste for annotations
- Annotation and label rotation
- New equations editor
- Commands to create and modify annotations
- Release date?
- Nightly builds available at:
https://cytoscape-builds.ucsd.edu/cytoscape-builds/Cytoscape-3.9.0/nightly/
Cytoscape Automation
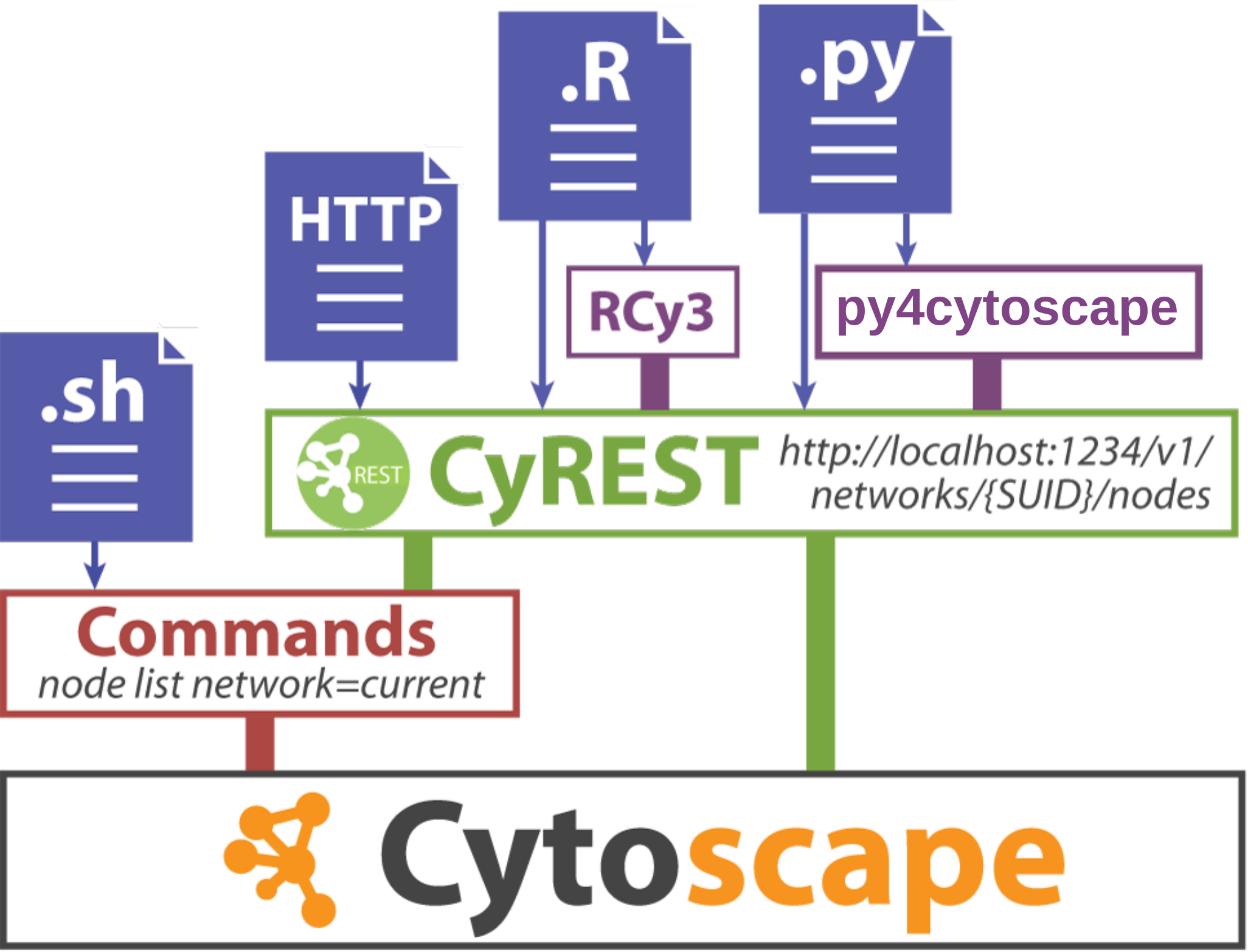
Cytoscape Automation
- RCy3
- 100's of methods to simply interaction between R scrips and Cytoscape, including:
- Create networks from igraph or graphNEL objects
- Load networks from Cytoscape into R
- Create and change styles
- Load tables from R data frames
- Load R data frames from Cytoscape
- Available via bioconductor
- py4cytoscape
- Generally, provides equivalent functionality as RCy3
- Available via pypy
