
Visualizing and Analysing Biological Networks with STRING and Cytoscape
Slides:
https://cytoscape.github.io/cytoscape-tutorials/presentations/cytoscape-and-string-2018-cpr.html
John "Scooter" Morris
Lars Juhl Jensen
Novo Nordisk Foundation Center for Protein Research
University of Copenhagen
15 November 2018
Goals and Motivations
- Understand the major applications of network biology
- Import your data into Cytoscape
- Master network layouts and data visualization
- Know where to find relevant Cytoscape apps and tutorials
- Understand STRING and how to integrate STRING data into Cytoscape
Introductions
- Executive director, Resource for Biocomputing, Visualization, and Informatics
- Roving Engineer, National Resource for Network Biology
- Cytoscape team since 2006
- Author of over a dozen Cytoscape apps
Introductions
- Group leader, Cellular Network Biology @ CPR
- Member of the STRING team since 2003
- PI of the COMPARTMENTS, TISSUES, and DISEASES databases
- Developer of the underlying text-mining software
Introductions
- Clinicians
- Bench Biologists
- Bioinformaticians
- Computer Scientists
- Chemists
- Mathematicians
- Other
Introductions
- None -- that's what I'm here for!
- Some network analysis, but what's Cytoscape?
- I've played with Cytoscape, but still have a lot to learn.
- I use Cytoscape all the time...
(hey, I could probably teach this course!)
Tour of Cytoscape
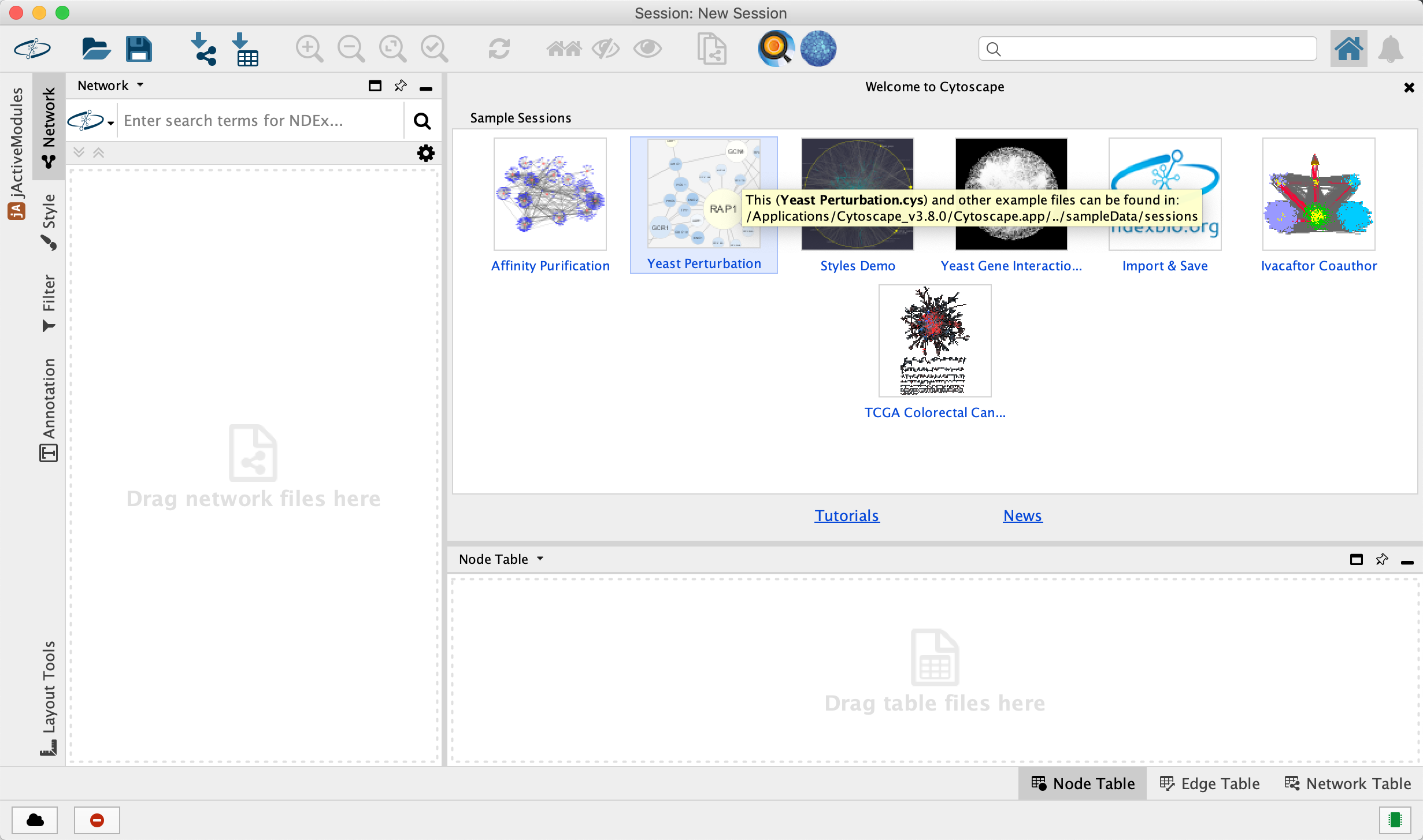
- Launch Cytoscape v3.7.0
Tour of Cytoscape
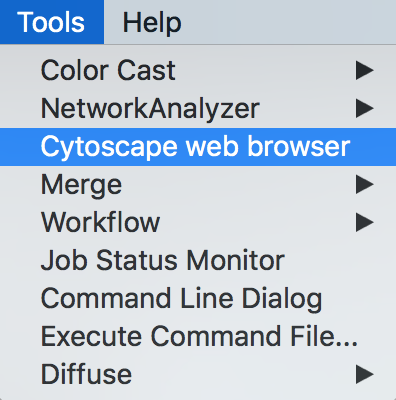
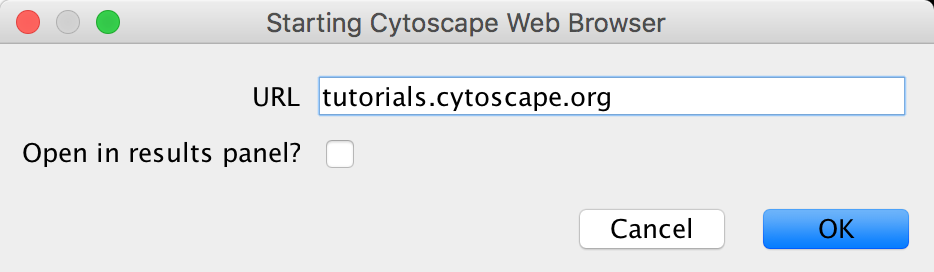
- Open CyBrowser
- Enter URL: tutorials.cytoscape.org
Tour of Cytoscape
- Click on Tour of Cytoscape
- Resize window to your preference
Tour of Cytoscape
- Loading networks
- Loading tables
- Selection and filtering
- Changing visual attributes
- Exporting images
- Saving sessions
Hands-on Exercises
Other tutorials are available at: tutorials.cytoscape.org
Introduction to STRING
Cytoscape & STRING
Wrap up
Questions and Discussion