
Data Analysis & Visualization in Cytoscape
Slides:
https://cytoscape.github.io/cytoscape-tutorials/presentations/embo-ppi-2018-rome.html
John "Scooter" Morris
Nadezhda Doncheva
Lars Juhl Jensen
Computational analysis of protein-protein interactions: Sequences, networks and diseases
4-19 October 2018, Rome Italy
Goals and Motivations
- Understand the major applications of network biology to the study of PPIs
- Find relevant networks and pathways
- Import your data into Cytoscape
- Master network layouts and data visualization
- Know where to find relevant Cytoscape apps and tutorials
Introductions
John "Scooter" Morris, UCSF
Nadezhda Doncheva, University of Copenhagen
Lars Juhl Jensen, University of Copenhagen
Introductions
- Clinicians
- Bench Biologists
- Bioinformaticians
- Computer Scientists
- Chemists
- Mathematicians
- Other
Introductions
- None -- that's what I'm here for!
- Some network analysis, but what's Cytoscape?
- I've played with Cytoscape, but still have a lot to learn.
- I use Cytoscape all the time...
(hey, I could probably teach this course!)
Tour of Cytoscape
- Launch Cytoscape v3.7.0
Tour of Cytoscape
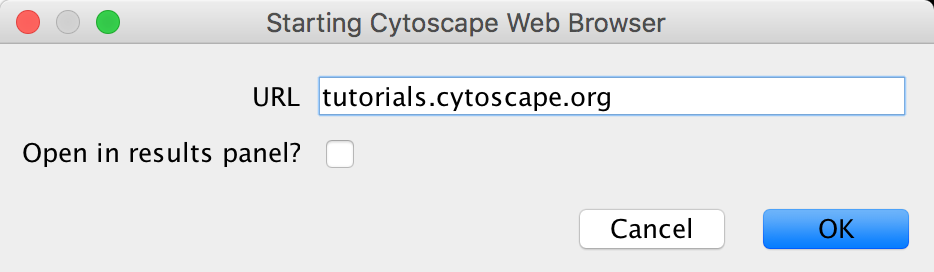
- Open CyBrowser
- Enter URL: tutorials.cytoscape.org
Tour of Cytoscape
- Click on Tour of Cytoscape
- Resize window to your preference
- Download either the Excel dataset or the tsv dataset.
- Loading networks
- Loading tables
- Selection and filtering
- Changing visual attributes
- Exporting images
- Saving sessions
Hands-on Exercises
Other tutorials are available at: tutorials.cytoscape.org
Wrap up
Questions and Discussion
"Exam"
To make sure you now have a working knowledge of Cytoscape, choose one of the following datasets to analyze in Cytoscape:
- Colins (2007): A merged dataset of three yeast APMS experiments. Find the putative complexes.