RNA-Seq Data Network Analysis
This protocol describes a network analysis workflow in Cytoscape for differentially expressed genes from an RNA-Seq experiment. Overall workflow:
- Finding a set of differentially expressed genes
- Retrieving relevant networks from public databases
- Integration and visualization of experimental data
- Network functional enrichment analysis
- Exporting network visualizations
Setup
- Install the stringApp from the Cytoscape App Store, or via
Apps → App Store → Show App Store .
Experimental Data
For this tutorial, we will use a dataset comparing transcriptomic differences between autistic and normal brain. The study has been published in Voineagu et al., and we will get a summarized dataset with fold change and p-value from the EBI Gene Expression Atlas.
- Download the data: Transcriptomic analysis of autistic brain reveals convergent molecular pathology.
- To open the tsv datafile in Excel, first launch Excel and open a blank workbook. Next, go to
Data → Get External Data → Import Text File... . - In the import wizard, select Delimited and in the next step select Tab.
- In the third step, you can select the Data Format for every column. The file has 4 columns of data: Gene ID, Gene Name, fold change and p-value. Make sure to change the format for the second column, Gene Name, to Text. You will have to scroll to the right to see the second column. Click Finish to complete the import.
Differentially Expressed Genes
We are going to define a set of up-regulated genes from the full dataset by filtering for fold change and p-value.
- Select the row containing data value headers (row 4) and select
Data → Filter . - In the drop-down for the fold change column, set a filter for fold change greater than 2. This should result in 263 genes.
- Next, one would normally filter out non-significant changes by filtering on the p-value as well, for example setting p-value less than 0.05. But in this case, all genes with a fold change greater than 2 already meet that cutoff.
- With the filter active, select and copy all entries in the Gene Name column.
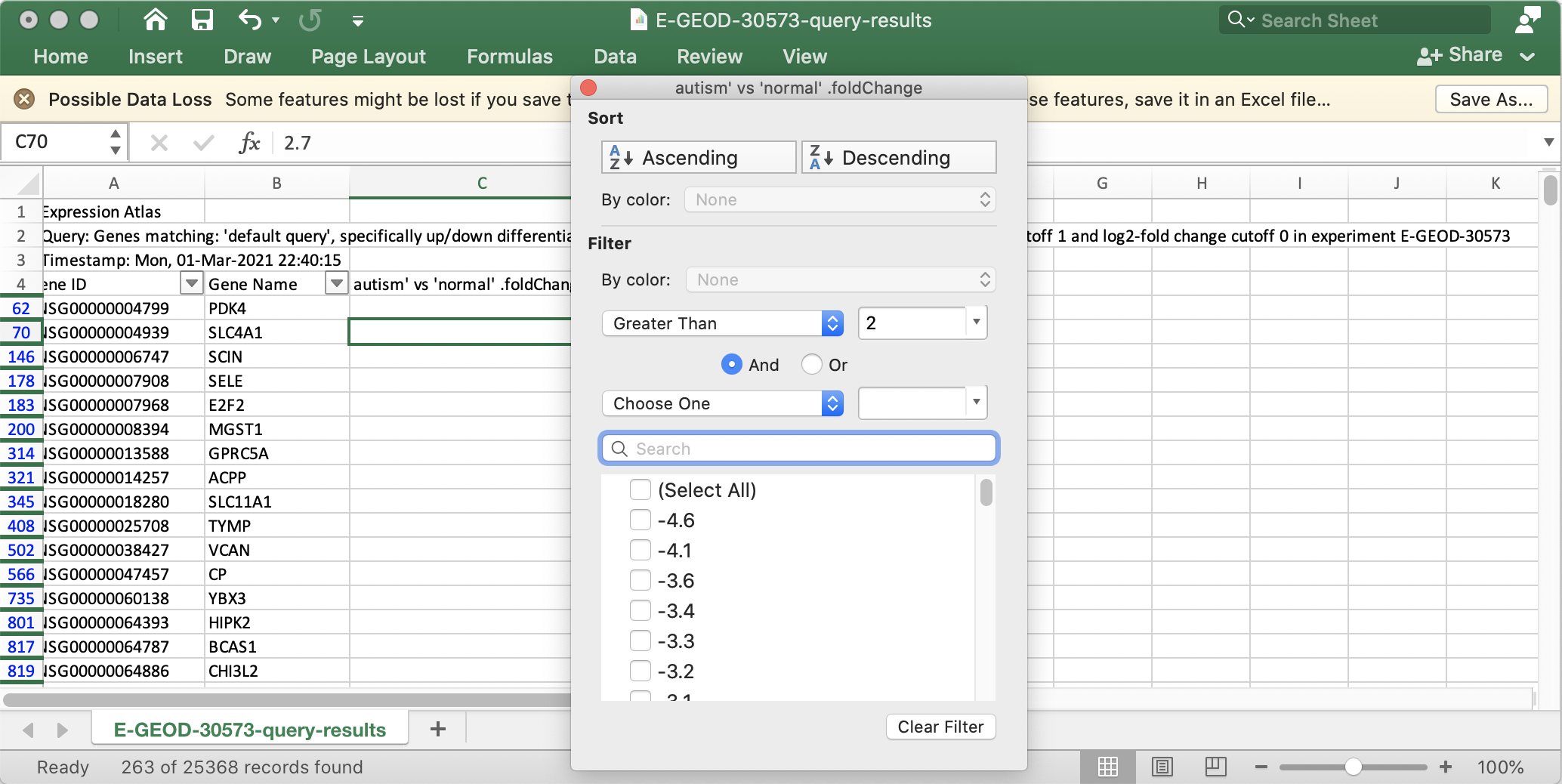
Retrieve Networks from STRING
To identify a relevant network, we will use the STRING database to find a network relevant to the list of up-regulated genes.
- Launch Cytoscape. In the
Network Search bar at the top of theNetwork Panel , selectSTRING protein query from the drop-down, and paste in the list of 263 up-regulated genes. - Open the options panel
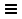 and confirm you are searching Homo sapiens with a Confidence cutoff of 0.40 and 0 Maximum additional interactors.
and confirm you are searching Homo sapiens with a Confidence cutoff of 0.40 and 0 Maximum additional interactors. - Click the search icon
 to search. If any of the search terms are ambiguous, a
to search. If any of the search terms are ambiguous, a Resolve Ambiguous Terms dialog will appear. ClickImport to continue with the import using the default choices. The resulting network will load automatically, and should have around 173 nodes.
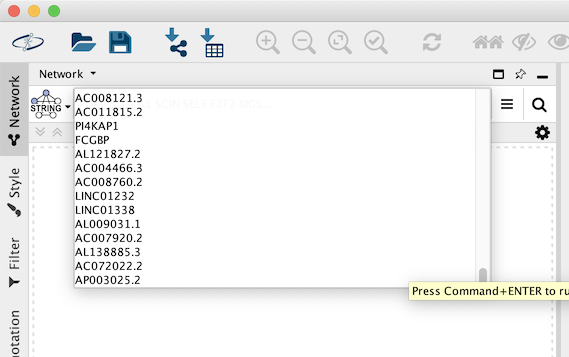
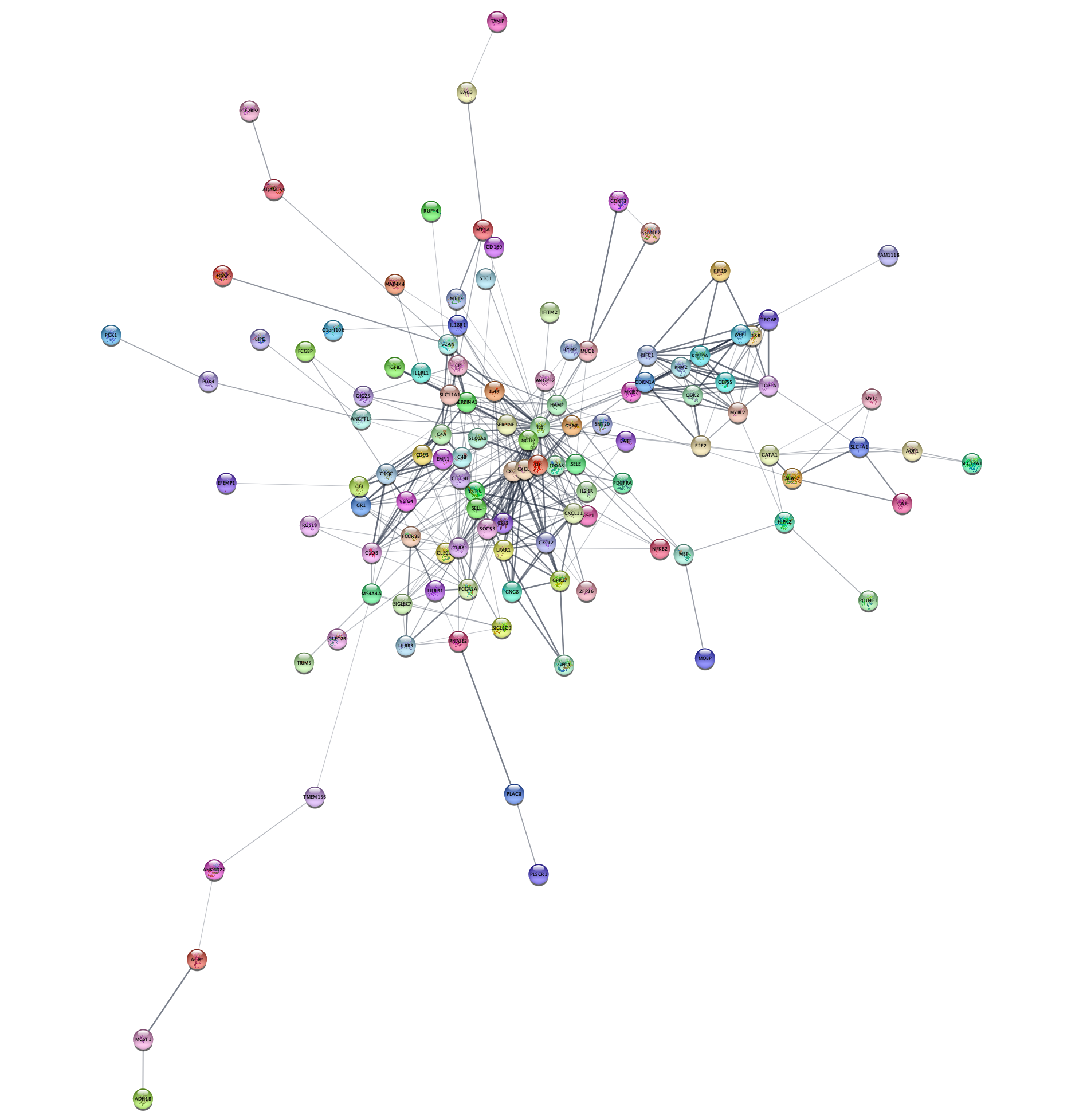
STRING Network Up-Regulated Genes
The resulting network contains up-regulated genes recognized by STRING, and interactions between them with an confidence score of 0.4 or greater.
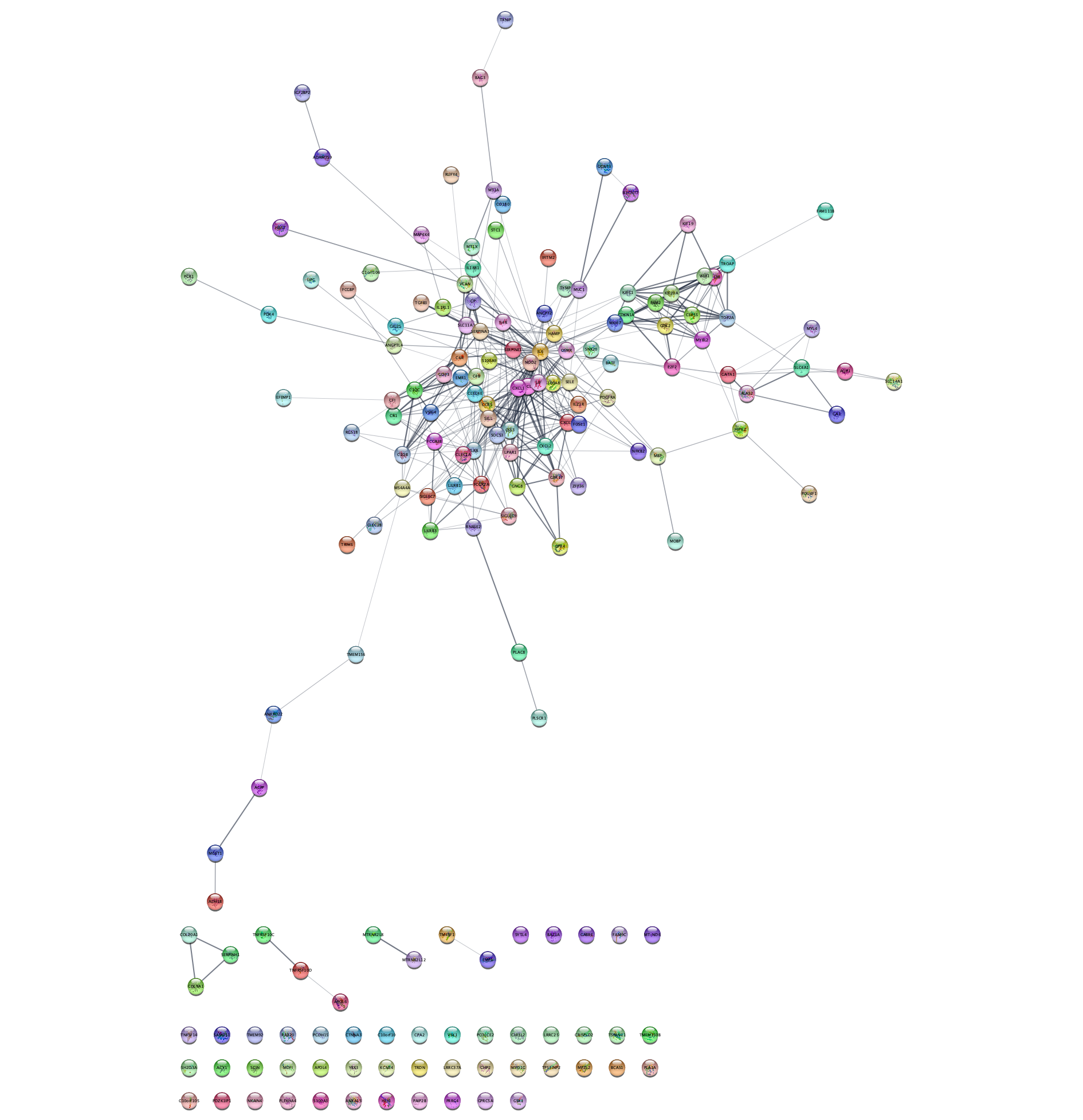
STRING Network Up-Regulated Genes
The networks consists of one large connected component, several smaller networks, and some unconnected nodes. We will use only the largest connected component for the rest of the tutorial.
- To select the largest connected component, select
Select → Nodes → Largest subnetwork . - Select
File → New Network → From Selected Nodes, All Edges .
Data Integration
Next we will import the RNA-Seq data and use them to create a visualization.
- Load the downloaded E-GEOD-30573-query-results.tsv file under
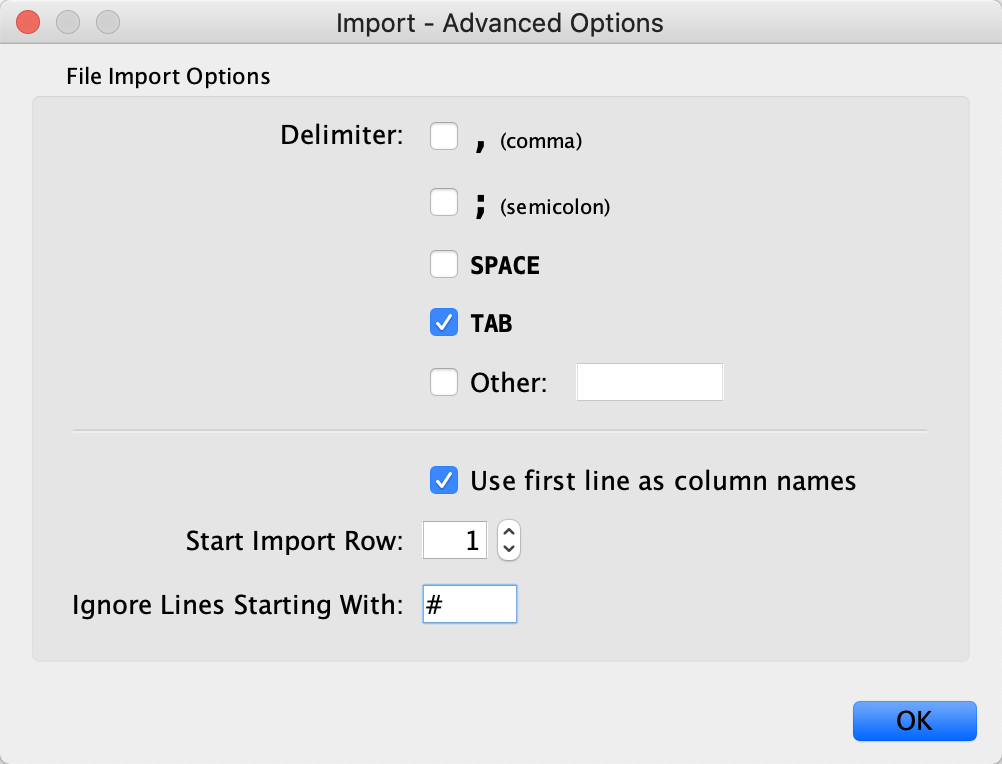
File menu by selectingImport → Table from File.... . Alternatively, drag and drop the data file directly onto theNode Table . - In
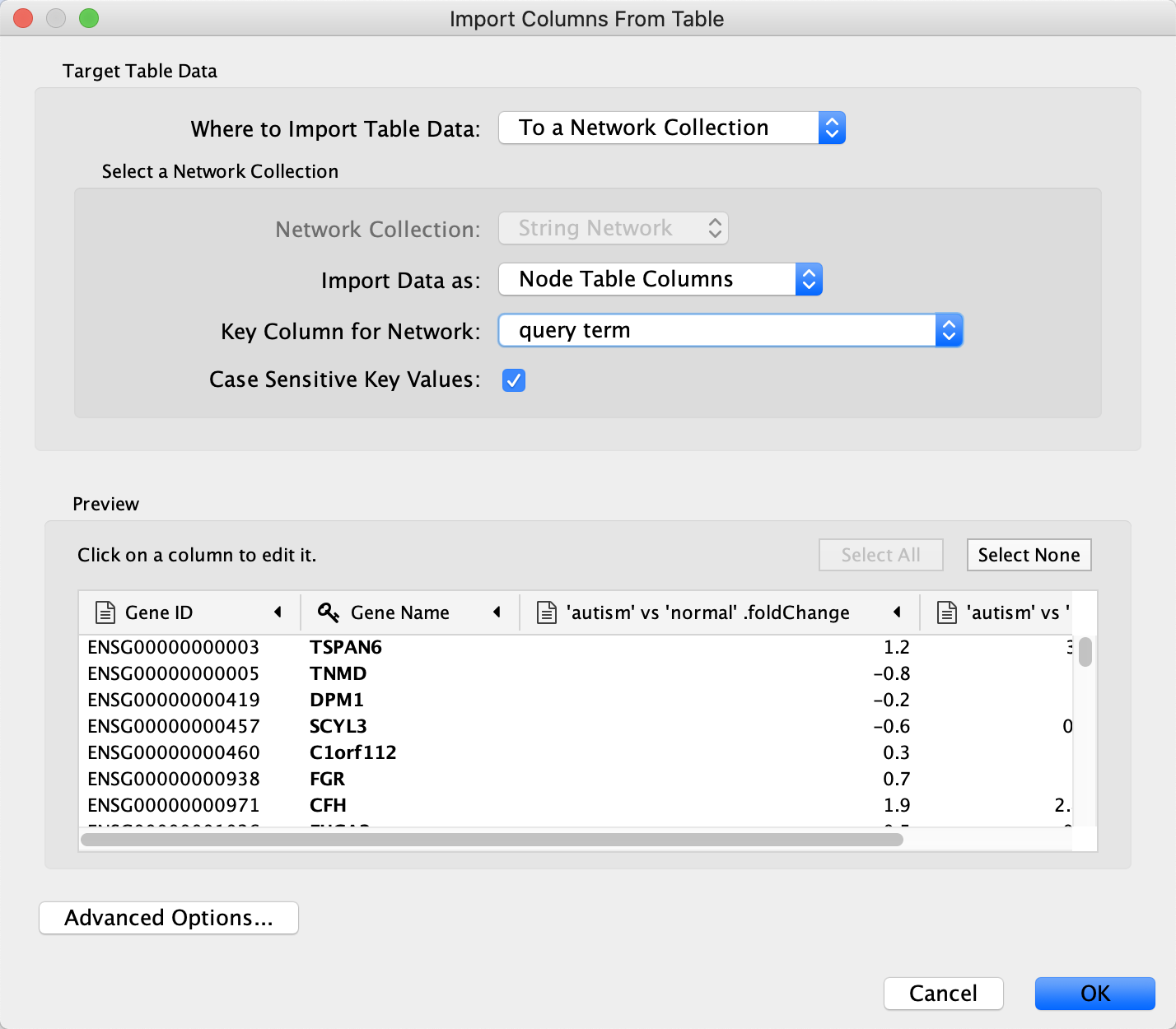
Advanced Options... , in theIgnore Lines Starting With field, enter #, to exclude the additional lines at the beginning of the data file. - Select the query term column as the Key column for Network and select the Gene Name column as the key column by clicking on the header and selecting the key symbol.
- Click
OK to import. Two new columns of data will be added to theNode Table .
Visualization
Next, we will create a visualization of the imported data on the network. For more detailed information on data visualization, see the Visualizing Data tutorial.
- In the
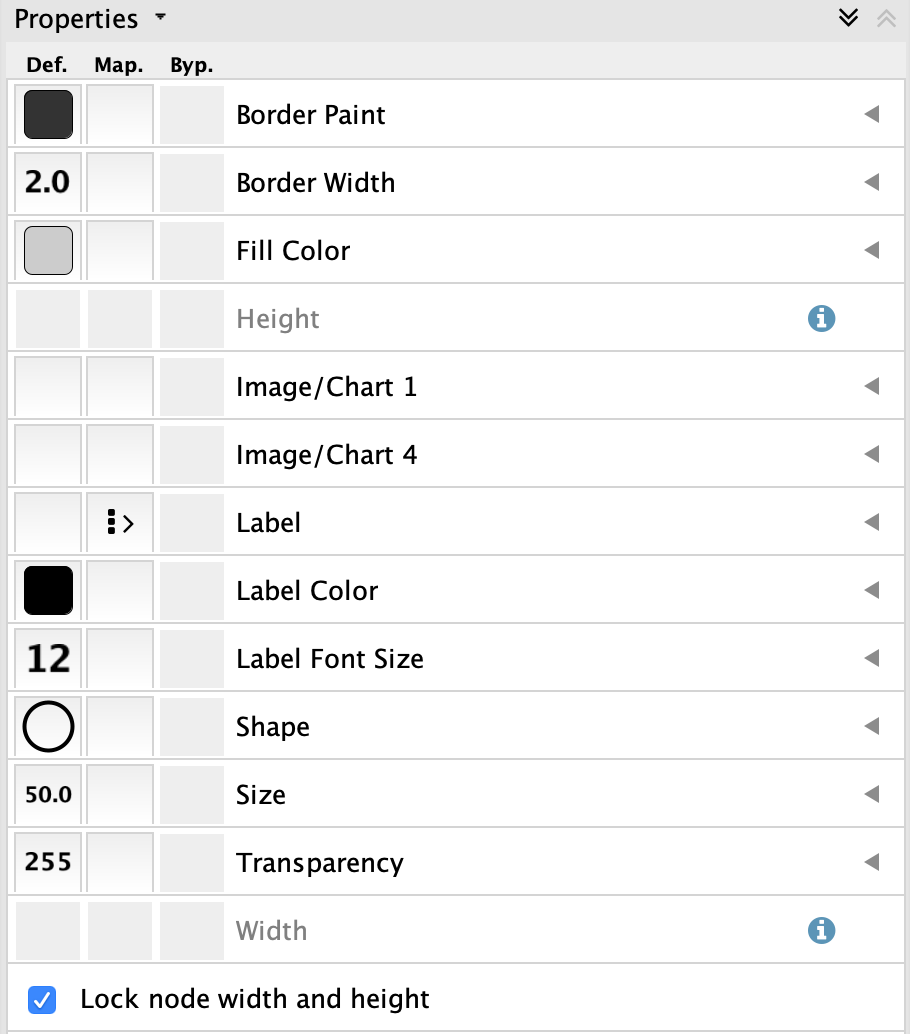
Style tab of theControl Panel , switch the style from STRING style to default in the drop-down at the top. - Change the default node
Shape to ellipse and checkLock node width and height . - Set the default node
Size to 50. - Set the default node
Fill Color to light gray. - Set the default
Border Width to 2, and make the defaultBorder Paint dark gray.
Visualization
- For node
Fill Color , create a continuous mapping for'autism' vs 'normal' .foldChange . - Double-cllick the color mapping to open the
Continuous Mapping Editor and click theCurrent Palette . Select the ColorBrewer yellow-orange-red shades gradient. - Finally, for node
Label , set a passthrough mapping fordisplay name . - Save your new visualization under
Copy Style... in theOptions menu of theStyle interface, and name it de genes up.
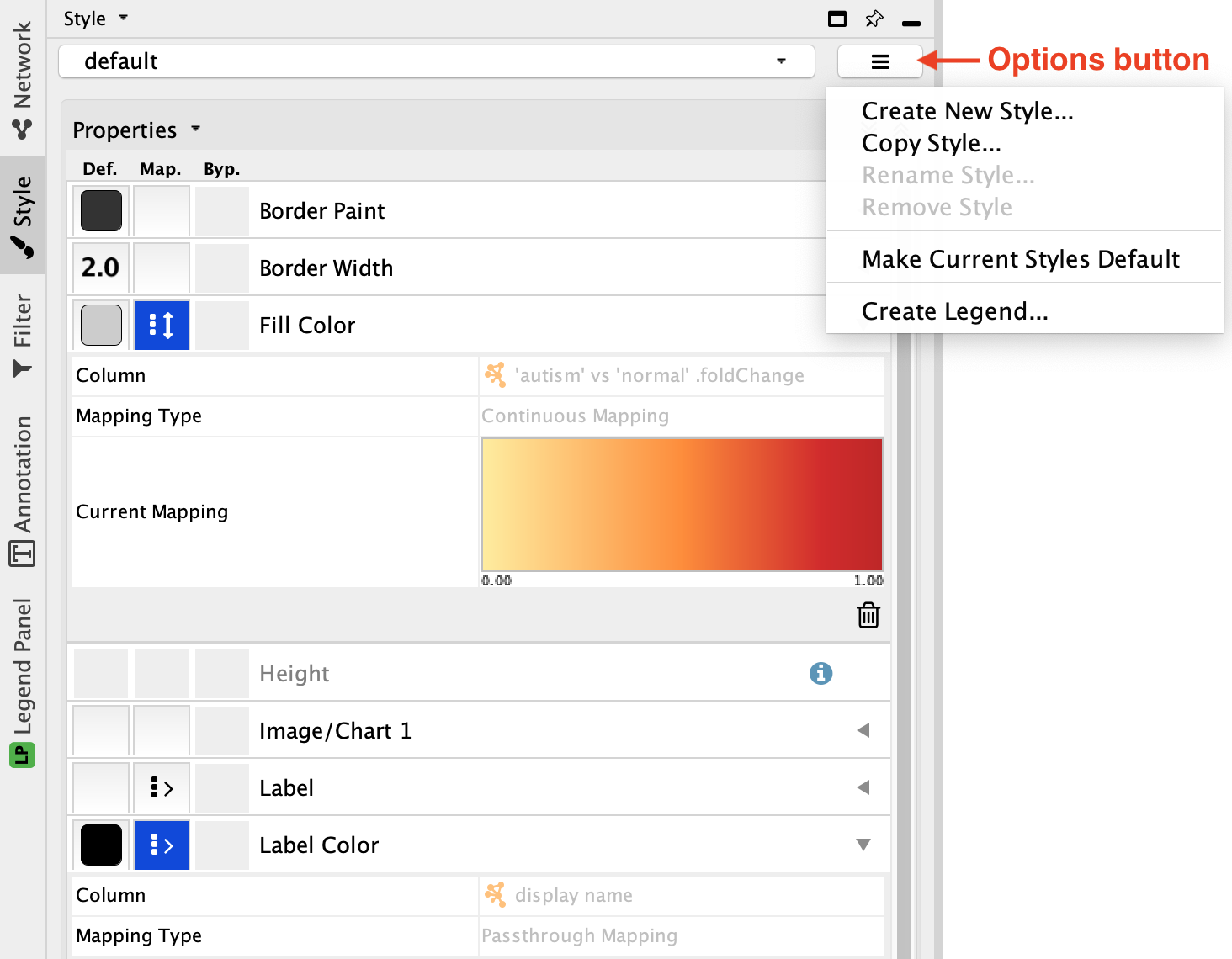
Visualization
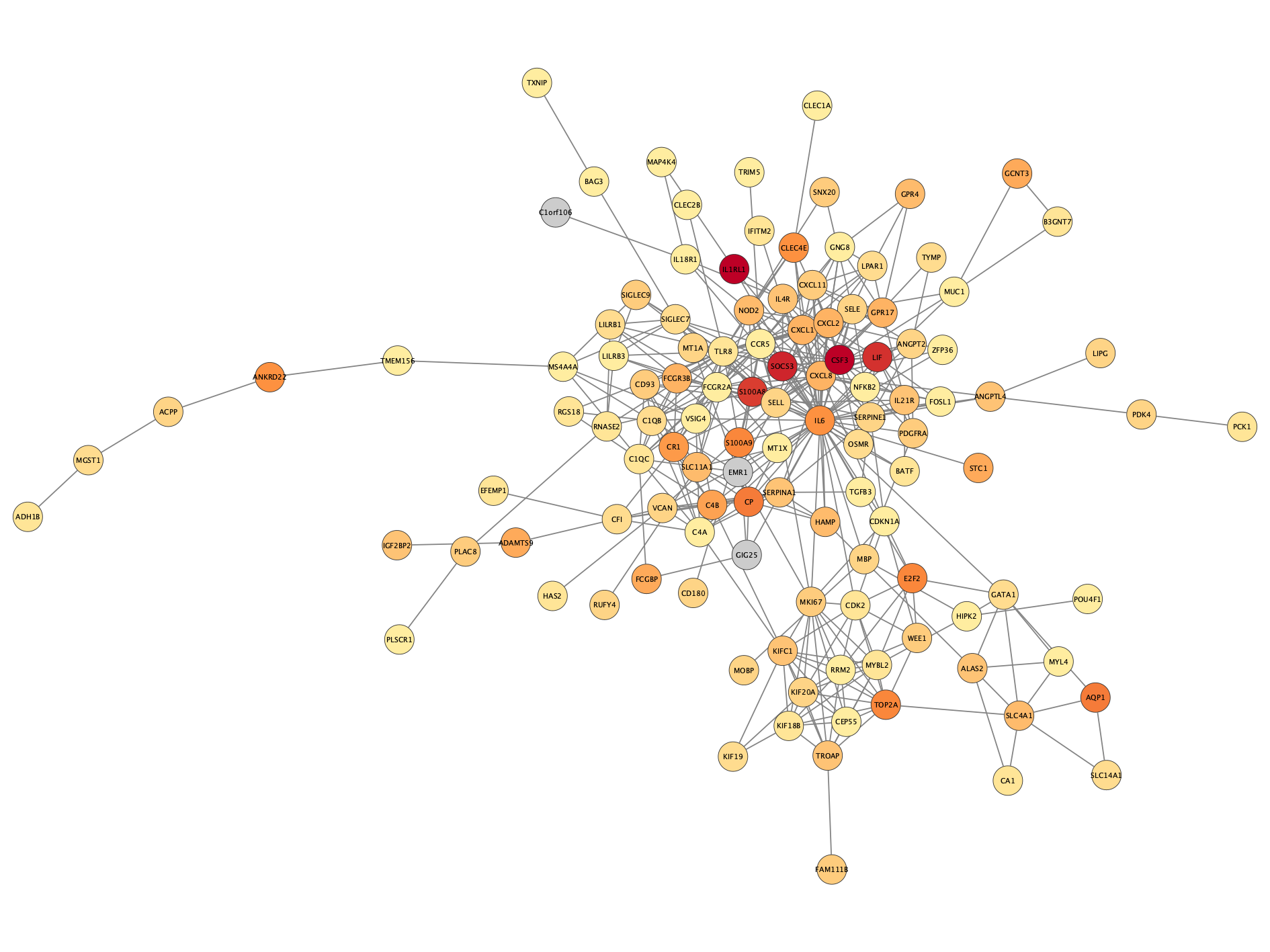
Apply the  button in the toolbar. The network will now look something like this:
button in the toolbar. The network will now look something like this:
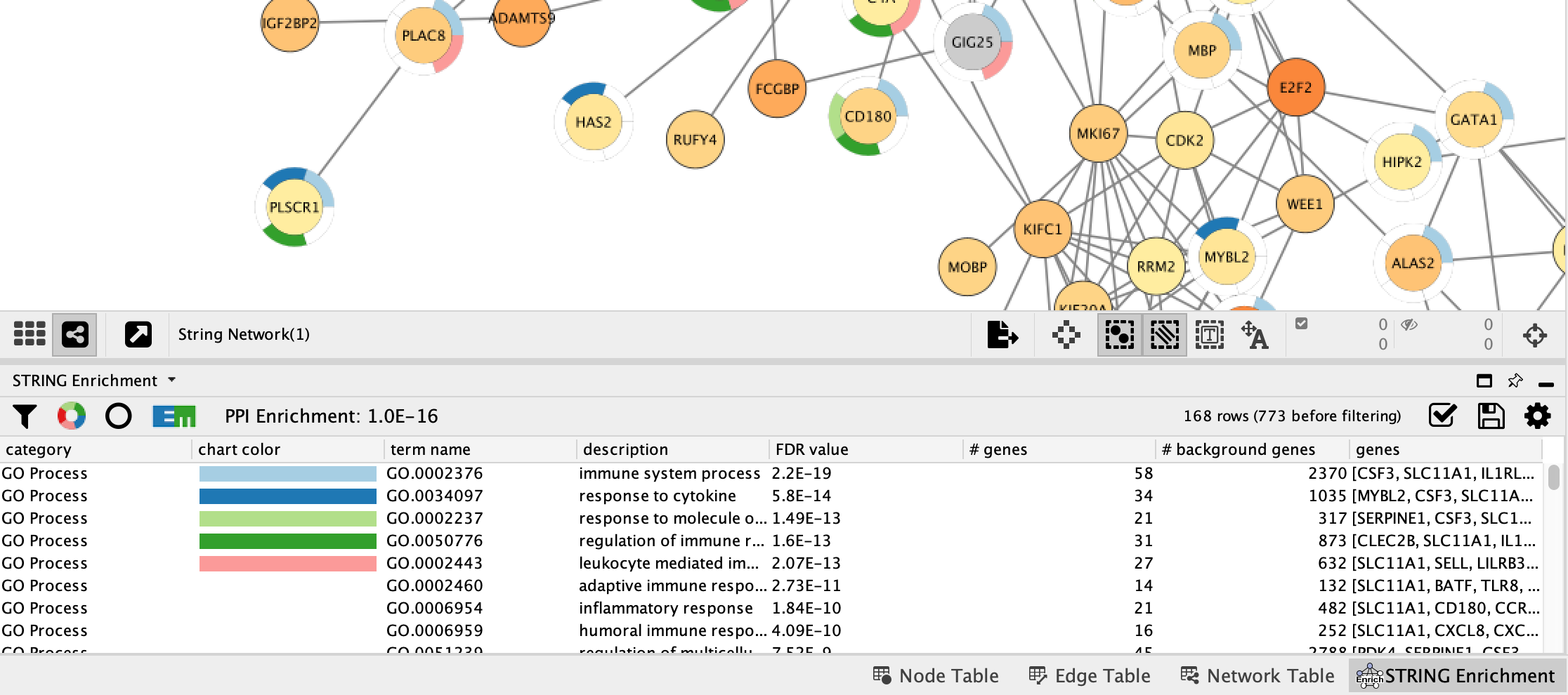
STRING Enrichment
The STRING app has built-in enrichment analysis functionality, which includes enrichment for Gene Ontology, InterPro, KEGG Pathways, and PFAM.
- In the STRING tab of the
Results Panel , click theFunctional Enrichment button. Keep the default settings.
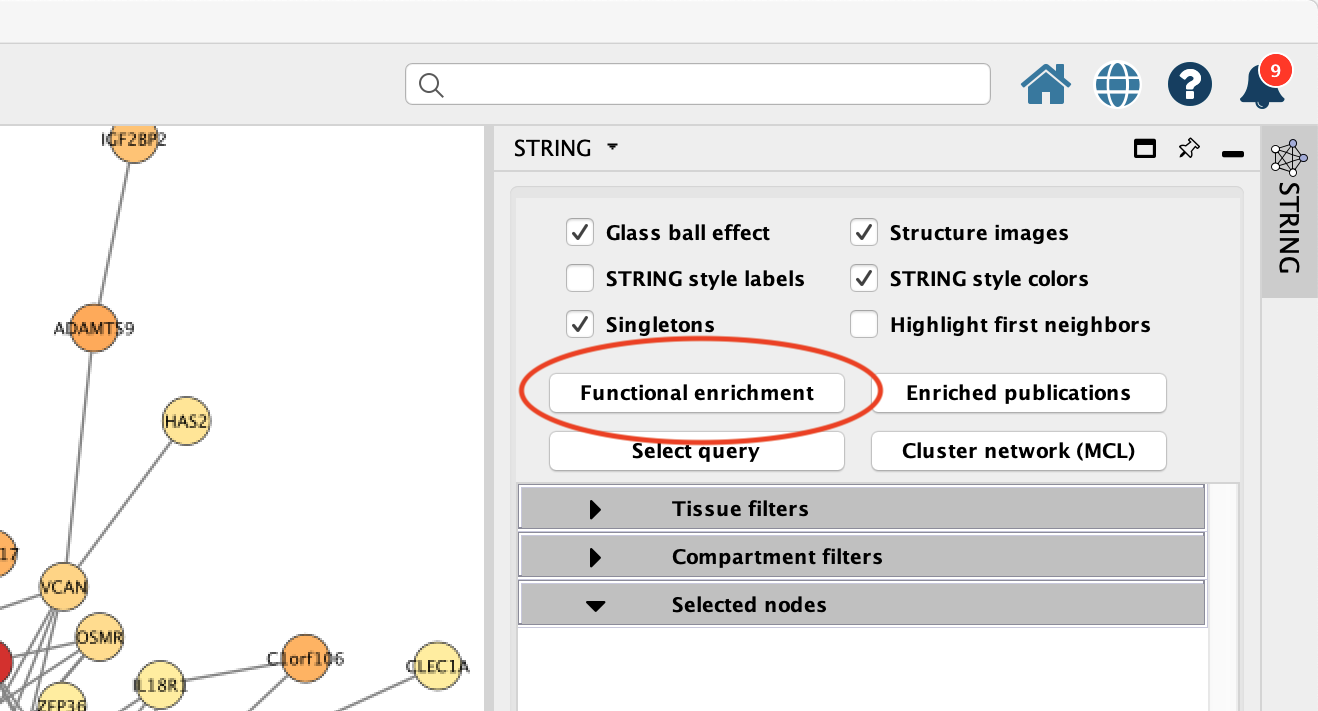
STRING Enrichment
- When the enrichment analysis is complete, a new tab titled
STRING Enrichment will open in theTable Panel .
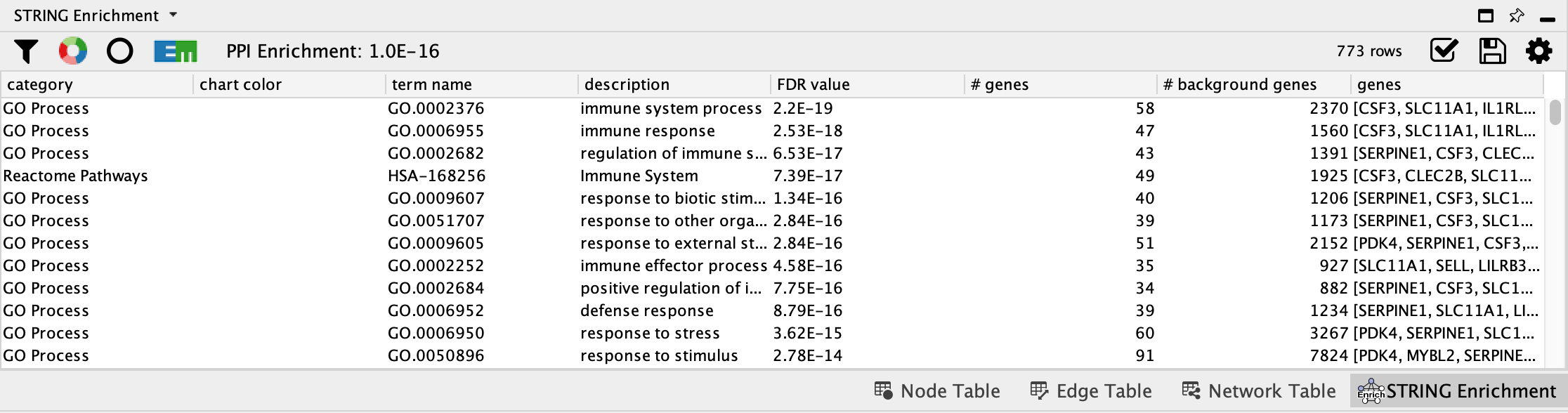
STRING Enrichment
The STRING app includes several options for filtering and displaying the enrichment results. The features are all available at the top of the
- At the top left of the STRING enrichment tab, click the filter icon
 . Select
. Select GO Biological Process and check theRemove redundant terms check-box. Then click OK. - Next, we will add a split donut chart to the nodes representing the top terms by clicking on
 .
. - Explore custom settings via
 in the top right of the STRING enrichment tab.
in the top right of the STRING enrichment tab.
Exporting Networks
Cytoscape provides a number of ways to save results and visualizations:
- As a session:
File → Save Session ,File → Save Session As... - As an image:
File → Export → Network to Image... - To the web:
File → Export → Network to Web Page... (Example) - To a public repository:
File → Export → Network to NDEx - As a graph format file:
File → Export → Network to File .
Formats:- CX JSON / CX2 JSON
- Cytoscape.js JSON
- GraphML
- PSI-MI
- XGMML
- SIF
