Introduction to Cytoscape:
Slides: https://cytoscape.org/cytoscape-tutorials/presentations/intro-cytoscape-2021-ismb.html#/
John "Scooter" Morris
ISMB/ECCB 2021 Tutorial: Reproducible omics data analysis workflows
July 22-23 2021
Goals and Motivations
- Understand the major applications of network biology
- Find relevant networks and pathways
- Import your data into Cytoscape
- Perform basic topological and other network analyses
- Perform network layouts and data visualization
- Know where to find relevant Cytoscape apps and tutorials
- Get started on using Cytoscape from R or Python
Introductions
- Executive director, Resource for Biocomputing, Visualization, and Informatics
- Roving Engineer, National Resource for Network Biology
- Cytoscape team since 2006
- Author of over two dozen Cytoscape apps
Caveats
- There are many more slides than we have time for
- Depending on your interests, we will skip entire sections
- The slides will continue to be available for your reference
Tour of Cytoscape
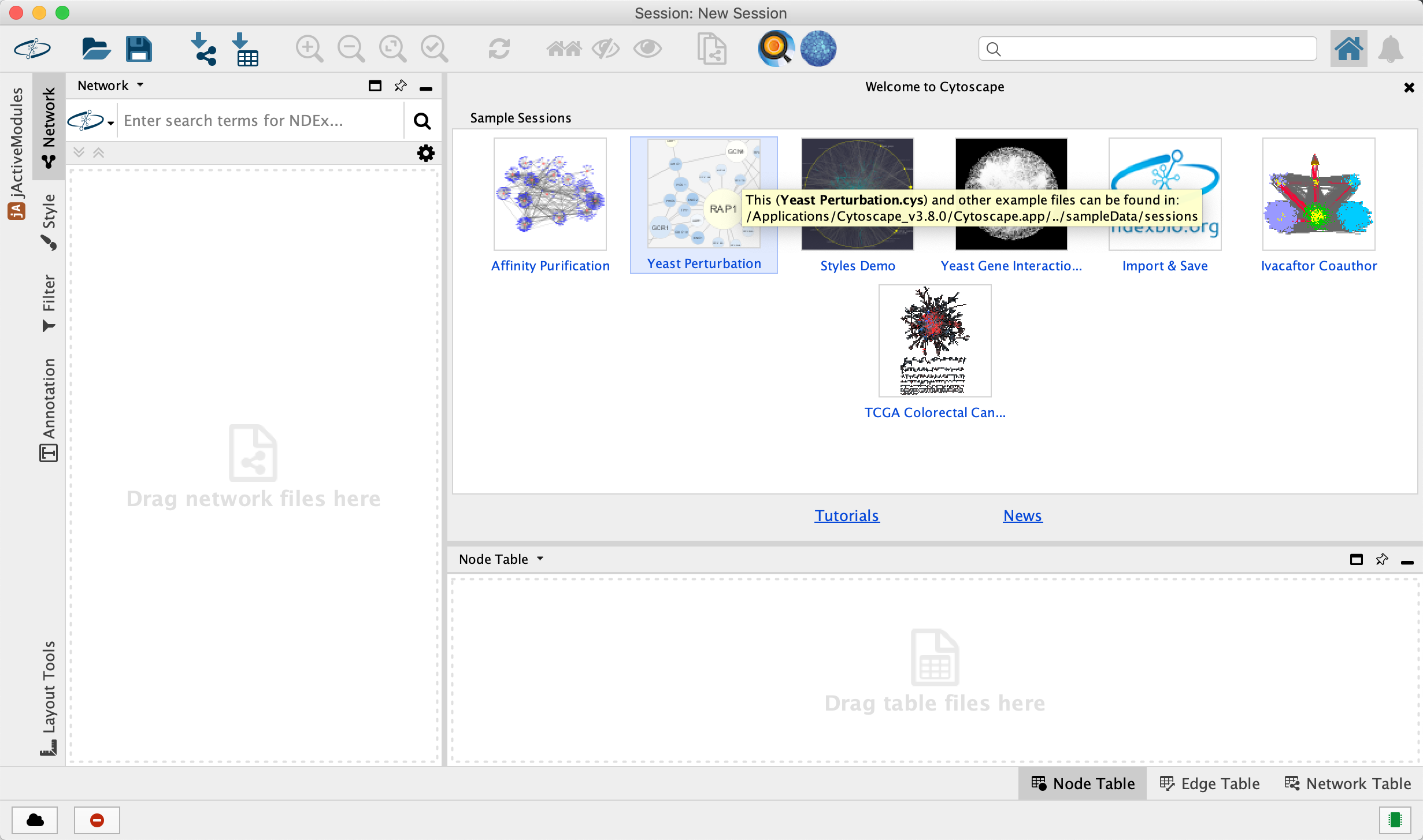
- Launch the latest version of Cytoscape
Tour of Cytoscape
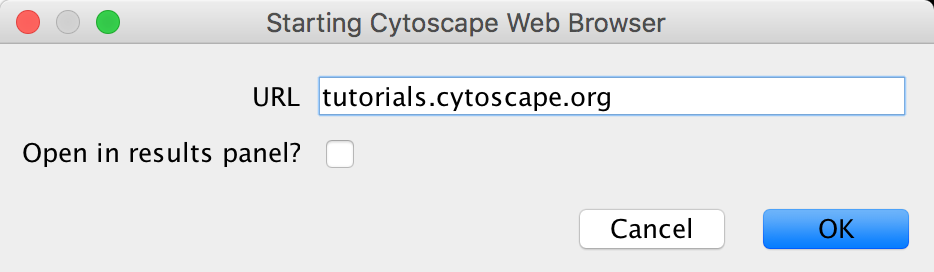
- Open CyBrowser
- Enter URL: tutorials.cytoscape.org
Tour of Cytoscape
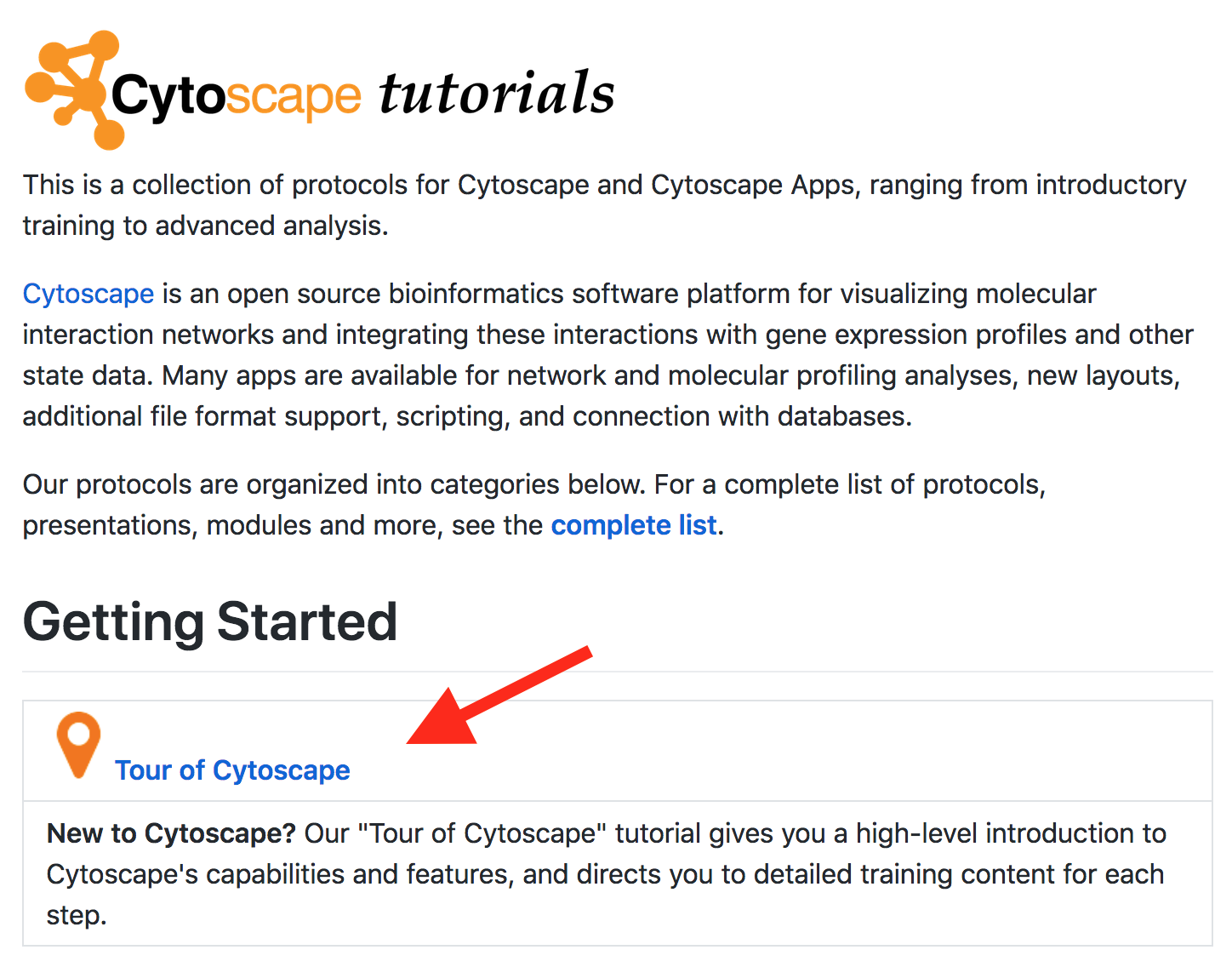
- Click on Tour of Cytoscape
- Resize window to your preference
Tour of Cytoscape
- Loading networks
- Loading tables
- Selection and filtering
- Changing visual attributes
- Exporting images
- Saving sessions
Hands-on Exercises
- Bulk RNA-Seq workflow: RNA-Seq Data Network Analysis
- Biological scenario: Basic Data Visualization
Hands-on Exercises
What have we learned?